Diff_0TO4_DEG_FULL = read.table("/Ets1/ProcessedData/Stage1_DEAdata.txt",sep = "\t", header=T,fill = TRUE)
Diff_0TO4_DEG_tmp = Diff_0TO4_DEG_FULL[which(Diff_0TO4_DEG_FULL[,7] <= 0.0001),]
Diff_0TO4_DEG_UP = Diff_0TO4_DEG_tmp[which(Diff_0TO4_DEG_tmp[,3] >= log2(1.5)),]
Diff_0TO4_DEG_DOWN = Diff_0TO4_DEG_tmp[which(Diff_0TO4_DEG_tmp[,3] <= -log2(1.5)),]
Diff_4TO7_DEG_FULL = read.table("/Ets1/ProcessedData/Stage2_DEAdata.txt",sep = "\t", header=T,fill = TRUE)
Diff_4TO7_DEG_tmp = Diff_4TO7_DEG_FULL[which(Diff_4TO7_DEG_FULL[,7] <= 0.0001),]
Diff_4TO7_DEG_UP = Diff_4TO7_DEG_tmp[which(Diff_4TO7_DEG_tmp[,3] >= log2(1.5)),]
Diff_4TO7_DEG_DOWN = Diff_4TO7_DEG_tmp[which(Diff_4TO7_DEG_tmp[,3] <= -log2(1.5)),]
Diff_7TO10_DEG_FULL = read.table("/Ets1/ProcessedData/Stage3_DEAdata.txt",sep = "\t", header=T,fill = TRUE)
Diff_7TO10_DEG_tmp = Diff_7TO10_DEG_FULL[which(Diff_7TO10_DEG_FULL[,7] <= 0.0001),]
Diff_7TO10_DEG_UP = Diff_7TO10_DEG_tmp[which(Diff_7TO10_DEG_tmp[,3] >= log2(1.5)),]
Diff_7TO10_DEG_DOWN = Diff_7TO10_DEG_tmp[which(Diff_7TO10_DEG_tmp[,3] <= -log2(1.5)),]
Diff_0TO4_DEG_UP_gene = Diff_0TO4_DEG_UP[,1]
Diff_4TO7_DEG_UP_gene = Diff_4TO7_DEG_UP[,1]
Diff_7TO10_DEG_UP_gene = Diff_7TO10_DEG_UP[,1]
Diff_0TO4_DEG_DOWN_gene = Diff_0TO4_DEG_DOWN[,1]
Diff_4TO7_DEG_DOWN_gene = Diff_4TO7_DEG_DOWN[,1]
Diff_7TO10_DEG_DOWN_gene = Diff_7TO10_DEG_DOWN[,1]
Diff_0TO4_DEG_UP_gene = as.data.frame(Diff_0TO4_DEG_UP_gene)
Diff_4TO7_DEG_UP_gene = as.data.frame(Diff_4TO7_DEG_UP_gene)
Diff_7TO10_DEG_UP_gene = as.data.frame(Diff_7TO10_DEG_UP_gene)
Diff_0TO4_DEG_DOWN_gene = as.data.frame(Diff_0TO4_DEG_DOWN_gene)
Diff_4TO7_DEG_DOWN_gene = as.data.frame(Diff_4TO7_DEG_DOWN_gene)
Diff_7TO10_DEG_DOWN_gene = as.data.frame(Diff_7TO10_DEG_DOWN_gene)
colnames(Diff_0TO4_DEG_UP_gene) = "gene"
colnames(Diff_4TO7_DEG_UP_gene) = "gene"
colnames(Diff_7TO10_DEG_UP_gene) = "gene"
colnames(Diff_0TO4_DEG_DOWN_gene) = "gene"
colnames(Diff_4TO7_DEG_DOWN_gene) = "gene"
colnames(Diff_7TO10_DEG_DOWN_gene) = "gene"
Diff_0TO4_DEG = rbind(Diff_0TO4_DEG_UP_gene,Diff_0TO4_DEG_DOWN_gene)
Diff_4TO7_DEG = rbind(Diff_4TO7_DEG_UP_gene,Diff_4TO7_DEG_DOWN_gene)
Diff_7TO10_DEG = rbind(Diff_7TO10_DEG_UP_gene,Diff_7TO10_DEG_DOWN_gene)
Diff_0TO4_DEG_PLOT = Diff_0TO4_DEG
Diff_4TO7_DEG_PLOT = Diff_4TO7_DEG
Diff_7TO10_DEG_PLOT = Diff_7TO10_DEG
Diff_0TO4_DEG_PLOT$S1 = 100
Diff_4TO7_DEG_PLOT$S2 = 10
Diff_7TO10_DEG_PLOT$S3 = 1
UPSET_PLOT = full_join(Diff_0TO4_DEG_PLOT,Diff_4TO7_DEG_PLOT,by="gene")
UPSET_PLOT = full_join(UPSET_PLOT,Diff_7TO10_DEG_PLOT,by="gene")
UPSET_PLOT[is.na(UPSET_PLOT)] = 0
UPSET_PLOT$score = UPSET_PLOT[,2]+UPSET_PLOT[,3]+UPSET_PLOT[,4]
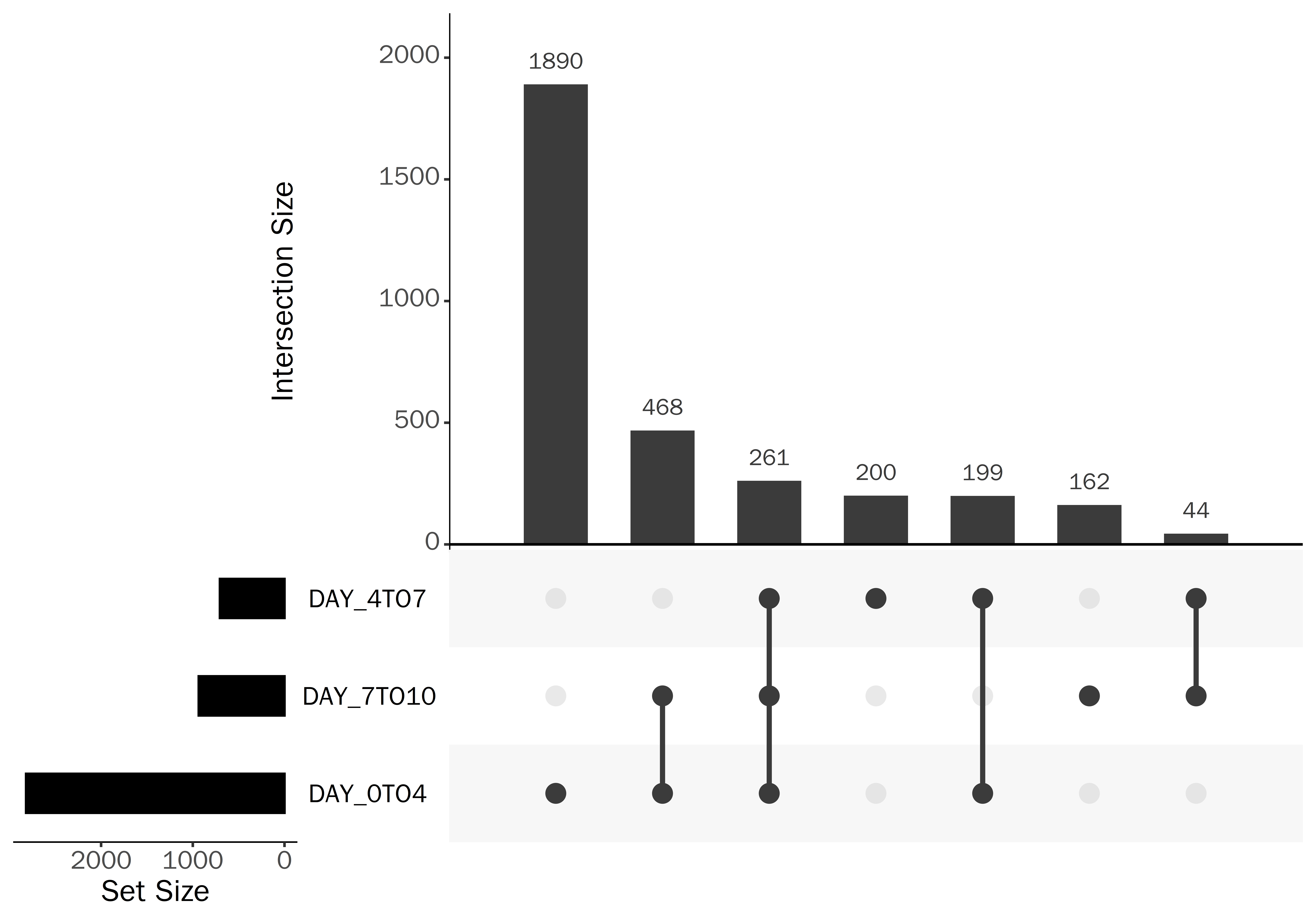
input <- c(
DAY_0TO4 = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 100),]),
DAY_4TO7 = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 10),]),
DAY_7TO10 = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 1),]),
"DAY_0TO4&DAY_4TO7&DAY_7TO10" = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 111),]),
"DAY_0TO4&DAY_4TO7" = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 110),]),
"DAY_0TO4&DAY_7TO10" = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 101),]),
"DAY_4TO7&DAY_7TO10" = nrow(UPSET_PLOT[which(UPSET_PLOT[,5] == 11),])
)
options(repr.plot.width = 10, repr.plot.height = 7, repr.plot.res = 1000, repr.plot.pointsize = 20)
upset(fromExpression(input),
nintersects = 19,
nsets = 4,
sets.bar.color = c("black"),
order.by = "freq",
decreasing = T,
mb.ratio = c(0.6, 0.4),
number.angles = 0,
text.scale = 2,
point.size = 5,
line.size = 1
)