Diff_0TO4_DEG_UP_GO = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/3_DEGs/DEG_result/Diff_0TO4_DEG_UP_GO.txt",sep = "\t", header=T,fill = TRUE)
Diff_4TO7_DEG_UP_GO = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/3_DEGs/DEG_result/Diff_4TO7_DEG_UP_GO.txt",sep = "\t", header=T,fill = TRUE)
Diff_7TO10_DEG_UP_GO = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/3_DEGs/DEG_result/Diff_7TO10_DEG_UP_GO.txt",sep = "\t", header=T,fill = TRUE)
Diff_0TO4_DEG_UP_GO = Diff_0TO4_DEG_UP_GO[,c(2,10)]
Diff_4TO7_DEG_UP_GO = Diff_4TO7_DEG_UP_GO[,c(2,10)]
Diff_7TO10_DEG_UP_GO = Diff_7TO10_DEG_UP_GO[,c(2,10)]
colnames(Diff_0TO4_DEG_UP_GO)[2] = "RF_0TO4"
colnames(Diff_4TO7_DEG_UP_GO)[2] = "RF_4TO7"
colnames(Diff_7TO10_DEG_UP_GO)[2] = "RF_7TO10"
Diff_DEG_UP_GO = full_join(Diff_0TO4_DEG_UP_GO,Diff_4TO7_DEG_UP_GO,by="Description")
Diff_DEG_UP_GO = full_join(Diff_DEG_UP_GO,Diff_7TO10_DEG_UP_GO,by="Description")
Diff_DEG_UP_GO[is.na(Diff_DEG_UP_GO)] = 0
Diff_DEG_UP_GO$label = "NOT"
for(i in 1:nrow(Diff_DEG_UP_GO)){
RF_0TO4 = Diff_DEG_UP_GO[i,2]
RF_4TO7 = Diff_DEG_UP_GO[i,3]
RF_7TO10 = Diff_DEG_UP_GO[i,4]
if(RF_0TO4 >= RF_4TO7){
if(RF_0TO4 >= RF_7TO10){
Diff_DEG_UP_GO[i,5] = "RF_0TO4"
}
}
if(RF_4TO7 >= RF_0TO4){
if(RF_4TO7 >= RF_7TO10){
Diff_DEG_UP_GO[i,5] = "RF_4TO7"
}
}
if(RF_7TO10 >= RF_0TO4){
if(RF_7TO10 >= RF_4TO7){
Diff_DEG_UP_GO[i,5] = "RF_7TO10"
}
}
}
Diff_DEG_UP_GO_RF_0TO4 = Diff_DEG_UP_GO[which(Diff_DEG_UP_GO[,5] == "RF_0TO4"),]
Diff_DEG_UP_GO_RF_4TO7 = Diff_DEG_UP_GO[which(Diff_DEG_UP_GO[,5] == "RF_4TO7"),]
Diff_DEG_UP_GO_RF_7TO10 = Diff_DEG_UP_GO[which(Diff_DEG_UP_GO[,5] == "RF_7TO10"),]
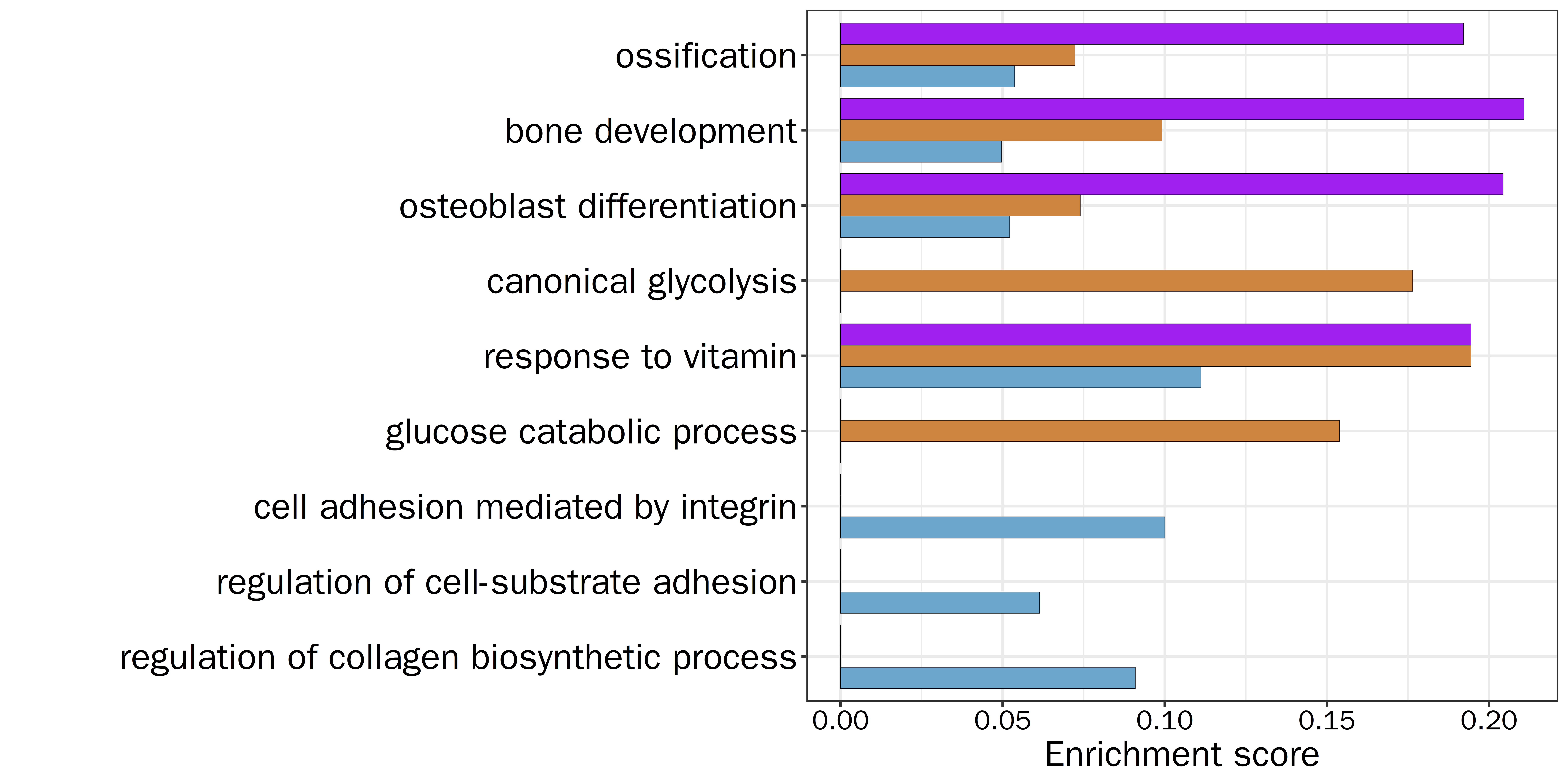
Diff_DEG_UP_GO_plot = rbind(Diff_DEG_UP_GO_RF_0TO4[which(Diff_DEG_UP_GO_RF_0TO4[,1] == "ossification"),],
Diff_DEG_UP_GO_RF_0TO4[which(Diff_DEG_UP_GO_RF_0TO4[,1] == "bone development"),],
Diff_DEG_UP_GO_RF_0TO4[which(Diff_DEG_UP_GO_RF_0TO4[,1] == "osteoblast differentiation"),],
Diff_DEG_UP_GO_RF_4TO7[which(Diff_DEG_UP_GO_RF_4TO7[,1] == "canonical glycolysis"),],
Diff_DEG_UP_GO_RF_4TO7[which(Diff_DEG_UP_GO_RF_4TO7[,1] == "response to vitamin"),],
Diff_DEG_UP_GO_RF_4TO7[which(Diff_DEG_UP_GO_RF_4TO7[,1] == "glucose catabolic process"),],
Diff_DEG_UP_GO_RF_7TO10[which(Diff_DEG_UP_GO_RF_7TO10[,1] == "cell adhesion mediated by integrin"),],
Diff_DEG_UP_GO_RF_7TO10[which(Diff_DEG_UP_GO_RF_7TO10[,1] == "regulation of cell-substrate adhesion"),],
Diff_DEG_UP_GO_RF_7TO10[which(Diff_DEG_UP_GO_RF_7TO10[,1] == "regulation of collagen biosynthetic process"),])
plot_DF = rbind(cbind(paste(9,Diff_DEG_UP_GO_plot[1,1],sep= " "),"3",Diff_DEG_UP_GO_plot[1,2]),
cbind(paste(9,Diff_DEG_UP_GO_plot[1,1],sep= " "),"2",Diff_DEG_UP_GO_plot[1,3]),
cbind(paste(9,Diff_DEG_UP_GO_plot[1,1],sep= " "),"1",Diff_DEG_UP_GO_plot[1,4]))
for(i in 2:nrow(Diff_DEG_UP_GO_plot)){
plot_DF_frag = rbind(cbind(paste(10-i,Diff_DEG_UP_GO_plot[i,1],sep= " "),"3",Diff_DEG_UP_GO_plot[i,2]),
cbind(paste(10-i,Diff_DEG_UP_GO_plot[i,1],sep= " "),"2",Diff_DEG_UP_GO_plot[i,3]),
cbind(paste(10-i,Diff_DEG_UP_GO_plot[i,1],sep= " "),"1",Diff_DEG_UP_GO_plot[i,4]))
plot_DF = rbind(plot_DF,plot_DF_frag)
}
plot_DF = as.data.frame(plot_DF)
colnames(plot_DF) = c("Description","cate","value")
plot_DF[,3] = as.numeric(plot_DF[,3])