1E
mm10_promoter_DB = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/DBs/mm10_promoter_2kb_DB.txt",sep = "\t", header=T,fill = TRUE)
mm10_promoter_DB = na.omit(mm10_promoter_DB)
mm10_promoter_DB = mm10_promoter_DB[which(mm10_promoter_DB[,13] <= 0.0001),]
mm10_promoter_DB_UP = mm10_promoter_DB[which(mm10_promoter_DB[,12] >= log2(1.5)),]
mm10_promoter_DB_DOWN = mm10_promoter_DB[which(mm10_promoter_DB[,12] <= -log2(1.5)),]
mm10_promoter_DB_UP = mm10_promoter_DB_UP[which(mm10_promoter_DB_UP[,9] >= 0),]
mm10_promoter_DB_UP = mm10_promoter_DB_UP[which(mm10_promoter_DB_UP[,10] >= 0),]
mm10_promoter_DB_DOWN = mm10_promoter_DB_DOWN[which(mm10_promoter_DB_DOWN[,9] <= 0),]
mm10_promoter_DB_DOWN = mm10_promoter_DB_DOWN[which(mm10_promoter_DB_DOWN[,10] <= 0),]
mm10_promoter_DB_UP_gene = mm10_promoter_DB_UP[,c(4,12)]
mm10_promoter_DB_DOWN_gene = mm10_promoter_DB_DOWN[,c(4,12)]
mm10_promoter_DB_UP_gene = unique(mm10_promoter_DB_UP_gene)
mm10_promoter_DB_DOWN_gene = unique(mm10_promoter_DB_DOWN_gene)
mm10_enhancer_DB = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/DBs/mm10_enhancer_DB.txt",sep = "\t", header=T,fill = TRUE)
mm10_enhancer_DB = na.omit(mm10_enhancer_DB)
mm10_enhancer_DB = mm10_enhancer_DB[which(mm10_enhancer_DB[,13] <= 0.0001),]
mm10_enhancer_DB_UP = mm10_enhancer_DB[which(mm10_enhancer_DB[,12] >= log2(1.5)),]
mm10_enhancer_DB_DOWN = mm10_enhancer_DB[which(mm10_enhancer_DB[,12] <= -log2(1.5)),]
mm10_enhancer_DB_UP = mm10_enhancer_DB_UP[which(mm10_enhancer_DB_UP[,9] >= 0),]
mm10_enhancer_DB_UP = mm10_enhancer_DB_UP[which(mm10_enhancer_DB_UP[,10] >= 0),]
mm10_enhancer_DB_DOWN = mm10_enhancer_DB_DOWN[which(mm10_enhancer_DB_DOWN[,9] <= 0),]
mm10_enhancer_DB_DOWN = mm10_enhancer_DB_DOWN[which(mm10_enhancer_DB_DOWN[,10] <= 0),]
mm10_enhancer_DB_UP_gene = mm10_enhancer_DB_UP[,c(4,12)]
mm10_enhancer_DB_DOWN_gene = mm10_enhancer_DB_DOWN[,c(4,12)]
mm10_enhancer_DB_UP_gene = unique(mm10_enhancer_DB_UP_gene)
mm10_enhancer_DB_DOWN_gene = unique(mm10_enhancer_DB_DOWN_gene)
mm10_promoter_2kb_DB = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/DBs/mm10_promoter_2kb_DB.txt", sep = "\t", header=F,fill = TRUE)
mm10_enhancer_DB = read.table("/data3/psg/NGS_2025/4DN/HiC_data/FiG1/DBs/mm10_enhancer_DB.txt", sep = "\t", header=F,fill = TRUE)
colnames(mm10_promoter_2kb_DB)[4] = "gene"
colnames(mm10_enhancer_DB)[4] = "gene"
mm10_promoter_DB_UP = inner_join(mm10_promoter_2kb_DB,mm10_promoter_DB_UP_gene,by="gene")
mm10_promoter_DB_DOWN = inner_join(mm10_promoter_2kb_DB,mm10_promoter_DB_DOWN_gene,by="gene")
mm10_enhancer_DB_UP = inner_join(mm10_enhancer_DB,mm10_enhancer_DB_UP_gene,by="gene")
mm10_enhancer_DB_DOWN = inner_join(mm10_enhancer_DB,mm10_enhancer_DB_DOWN_gene,by="gene")
mm10_promoter_DB_UP = mm10_promoter_DB_UP[,-12]
mm10_promoter_DB_DOWN = mm10_promoter_DB_DOWN[,-12]
mm10_enhancer_DB_UP = mm10_enhancer_DB_UP[,-12]
mm10_enhancer_DB_DOWN = mm10_enhancer_DB_DOWN[,-12]
mm10RAWdata = genes(TxDb.Mmusculus.UCSC.mm10.knownGene)
mm10RAWdata = as.data.frame(mm10RAWdata)
rownames(mm10RAWdata) = NULL
colnames(mm10RAWdata) = c("chrmosome","start","end","width","strand","ENTREZID")
mm10RAWdata_frag = AnnotationDbi::select(org.Mm.eg.db, keys=mm10RAWdata[,6], columns="SYMBOL", keytype="ENTREZID")
mm10RAWdata_tmp = inner_join(mm10RAWdata,mm10RAWdata_frag,by = "ENTREZID")
mm10data = mm10RAWdata_tmp[,c(7,1,2,3,4,5)]
mm10data = mm10data[!grepl("^.*Rik+|^Gm[0-9]+$|^AC[0-9]+.[0-9]$|^BC[0-9]+|ps-|-ps",mm10data$SYMBOL), ]
colnames(mm10data) = c("gene","gene_chrmosome","gene_start","gene_end","gene_width","gene_strand")
mm10data_strand = mm10data[,c(1,6)]
mm10_promoter_DB_UP = inner_join(mm10_promoter_DB_UP,mm10data_strand, by="gene")
mm10_enhancer_DB_UP = inner_join(mm10_enhancer_DB_UP,mm10data_strand, by="gene")
mm10_promoter_DB_DOWN = inner_join(mm10_promoter_DB_DOWN,mm10data_strand, by="gene")
mm10_enhancer_DB_DOWN = inner_join(mm10_enhancer_DB_DOWN,mm10data_strand, by="gene")
mm10_promoter_DB_UP[,2] = as.numeric(mm10_promoter_DB_UP[,2])
mm10_promoter_DB_UP[,3] = as.numeric(mm10_promoter_DB_UP[,3])
mm10_promoter_DB_DOWN[,2] = as.numeric(mm10_promoter_DB_DOWN[,2])
mm10_promoter_DB_DOWN[,3] = as.numeric(mm10_promoter_DB_DOWN[,3])
mm10_promoter_DB_UP[,9] = as.numeric(mm10_promoter_DB_UP[,9])
mm10_promoter_DB_UP[,10] = as.numeric(mm10_promoter_DB_UP[,10])
mm10_promoter_DB_DOWN[,9] = as.numeric(mm10_promoter_DB_DOWN[,9])
mm10_promoter_DB_DOWN[,10] = as.numeric(mm10_promoter_DB_DOWN[,10])
mm10_enhancer_DB_UP[,2] = as.numeric(mm10_enhancer_DB_UP[,2])
mm10_enhancer_DB_UP[,3] = as.numeric(mm10_enhancer_DB_UP[,3])
mm10_enhancer_DB_DOWN[,2] = as.numeric(mm10_enhancer_DB_DOWN[,2])
mm10_enhancer_DB_DOWN[,3] = as.numeric(mm10_enhancer_DB_DOWN[,3])
mm10_enhancer_DB_UP[,9] = as.numeric(mm10_enhancer_DB_UP[,9])
mm10_enhancer_DB_UP[,10] = as.numeric(mm10_enhancer_DB_UP[,10])
mm10_enhancer_DB_DOWN[,9] = as.numeric(mm10_enhancer_DB_DOWN[,9])
mm10_enhancer_DB_DOWN[,10] = as.numeric(mm10_enhancer_DB_DOWN[,10])
mm10_promoter_DB_UP$FCmean = (mm10_promoter_DB_UP[,9]+mm10_promoter_DB_UP[,10])/2
mm10_promoter_DB_DOWN$FCmean = (mm10_promoter_DB_DOWN[,9]+mm10_promoter_DB_DOWN[,10])/2
mm10_enhancer_DB_UP$FCmean = (mm10_enhancer_DB_UP[,9]+mm10_enhancer_DB_UP[,10])/2
mm10_enhancer_DB_DOWN$FCmean = (mm10_enhancer_DB_DOWN[,9]+mm10_enhancer_DB_DOWN[,10])/2
frag = mm10_enhancer_DB_UP[which(mm10_enhancer_DB_UP[,4] == mm10_enhancer_DB_UP_gene[1,1]),]
frag = frag[order(-frag[,15]),]
mm10_enhancer_DB_UP_final = frag[1,]
for(i in 2:nrow(mm10_enhancer_DB_UP_gene)){
frag = mm10_enhancer_DB_UP[which(mm10_enhancer_DB_UP[,4] == mm10_enhancer_DB_UP_gene[i,1]),]
frag = frag[order(-frag[,15]),]
mm10_enhancer_DB_UP_final_frag = frag[1,]
mm10_enhancer_DB_UP_final = rbind(mm10_enhancer_DB_UP_final,mm10_enhancer_DB_UP_final_frag)
}
frag = mm10_enhancer_DB_DOWN[which(mm10_enhancer_DB_DOWN[,4] == mm10_enhancer_DB_DOWN_gene[1,1]),]
frag = frag[order(frag[,15]),]
mm10_enhancer_DB_DOWN_final = frag[1,]
for(i in 2:nrow(mm10_enhancer_DB_DOWN_gene)){
frag = mm10_enhancer_DB_DOWN[which(mm10_enhancer_DB_DOWN[,4] == mm10_enhancer_DB_DOWN_gene[i,1]),]
frag = frag[order(frag[,15]),]
mm10_enhancer_DB_DOWN_final_frag = frag[1,]
mm10_enhancer_DB_DOWN_final = rbind(mm10_enhancer_DB_DOWN_final,mm10_enhancer_DB_DOWN_final_frag)
}
mm10_enhancer_DB_UP = mm10_enhancer_DB_UP_final
mm10_enhancer_DB_DOWN = mm10_enhancer_DB_DOWN_final
mm10_promoter_DB_UP_top100 = mm10_promoter_DB_UP[order(-mm10_promoter_DB_UP[,15]),][1:100,]
mm10_enhancer_DB_UP_top100 = mm10_enhancer_DB_UP[order(-mm10_enhancer_DB_UP[,15]),][1:100,]
mm10_promoter_DB_DOWN_top100 = mm10_promoter_DB_DOWN[order(mm10_promoter_DB_DOWN[,15]),][1:100,]
mm10_enhancer_DB_DOWN_top100 = mm10_enhancer_DB_DOWN[order(mm10_enhancer_DB_DOWN[,15]),][1:100,]
options(repr.plot.width = 5, repr.plot.height = 5, repr.plot.res = 1000, repr.plot.pointsize = 10)
data = rbind(mm10_promoter_DB_UP_top100,mm10_promoter_DB_DOWN_top100)
gene = data[,4]
ENTREZID = bitr(gene, fromType="SYMBOL", toType="ENTREZID", OrgDb="org.Mm.eg.db")
colnames(ENTREZID)=c("gene","ENTREZID")
data_addID = inner_join(data,ENTREZID,by = "gene")
df = data_addID
original_gene_list <- df$log2FoldChange
names(original_gene_list) <- as.character(df$ENTREZID)
gene_list<-na.omit(original_gene_list)
gene_list = sort(gene_list, decreasing = TRUE)
msigdbr_GOBP = msigdbr(species = "mouse", category = "C5", subcategory = "GO:BP") %>%
dplyr::select(gs_name, entrez_gene)
GSEA_result <- GSEA(gene_list,
TERM2GENE = msigdbr_GOBP,
pAdjustMethod = "BH",
seed=TRUE,
by = "fgsea",
pvalueCutoff = 1)
GSEA_result_pcut = GSEA_result[which(GSEA_result[,6] <= 0.05),2:9]
rownames(GSEA_result_pcut) = NULL
GSEA_result_pcut = GSEA_result_pcut[order(GSEA_result_pcut$enrichmentScore),]
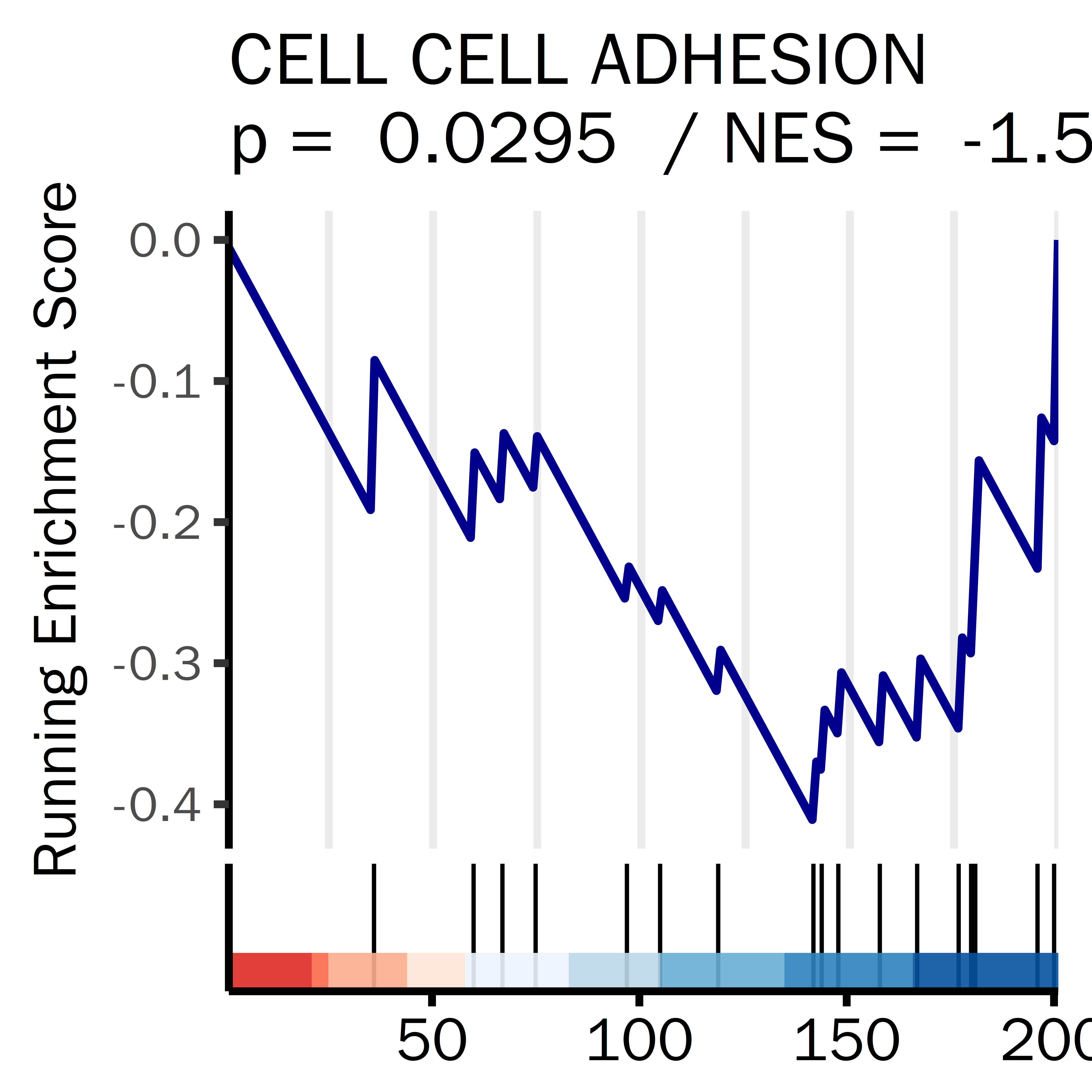
PLPLPLP = GSEA_result_pcut[grepl("GOBP_OSSIFICATION",GSEA_result_pcut[,1]),]
datassssset = PLPLPLP
datassssset = datassssset[1,]
name1 = "OSSIFICATION"
i = 1
ID = datassssset[i,1]
gseaplot2(GSEA_result,
rel_heights = c(0.5, 0.1, 0.1),
geneSetID = ID,
base_size = 20,
color = "darkred",
title =paste(name1,
"\np = ",
round(datassssset[i,5],4),
" / NES = ",
round(datassssset[i,4],4)),
subplots = 1:2,
ES_geom = "line")
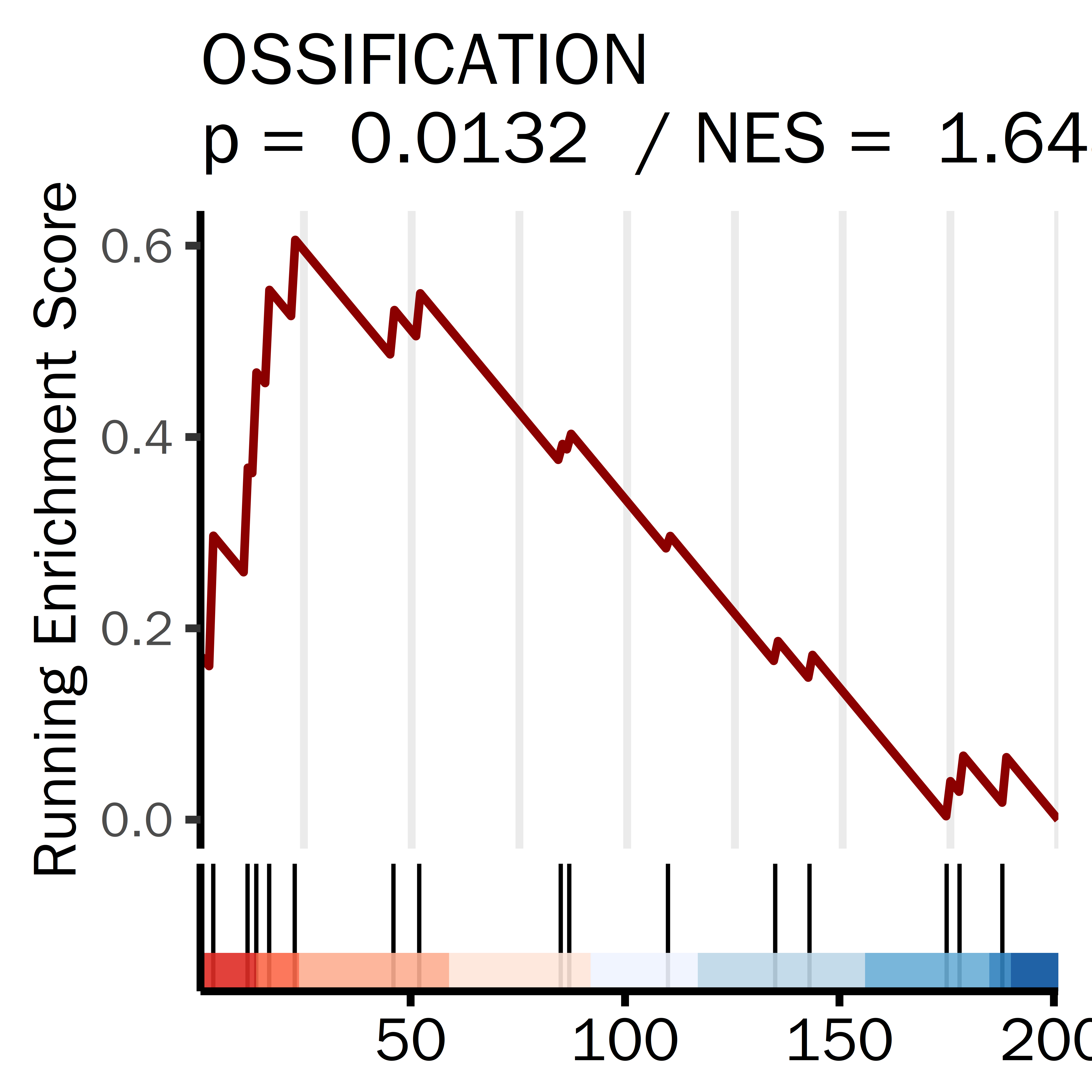
PLPLPLP = GSEA_result_pcut[grepl("GOBP_SKELETAL_SYSTEM_DEVELOPMENT",GSEA_result_pcut[,1]),]
datassssset = PLPLPLP
datassssset = datassssset[1,]
name1 = "SKELETAL SYSTEM DEVELOPMENT"
i = 1
ID = datassssset[i,1]
gseaplot2(GSEA_result,
rel_heights = c(0.5, 0.1, 0.1),
geneSetID = ID,
base_size = 20,
color = "darkred",
title =paste(name1,
"\np = ",
round(datassssset[i,5],4),
" / NES = ",
round(datassssset[i,4],4)),
subplots = 1:2,
ES_geom = "line")
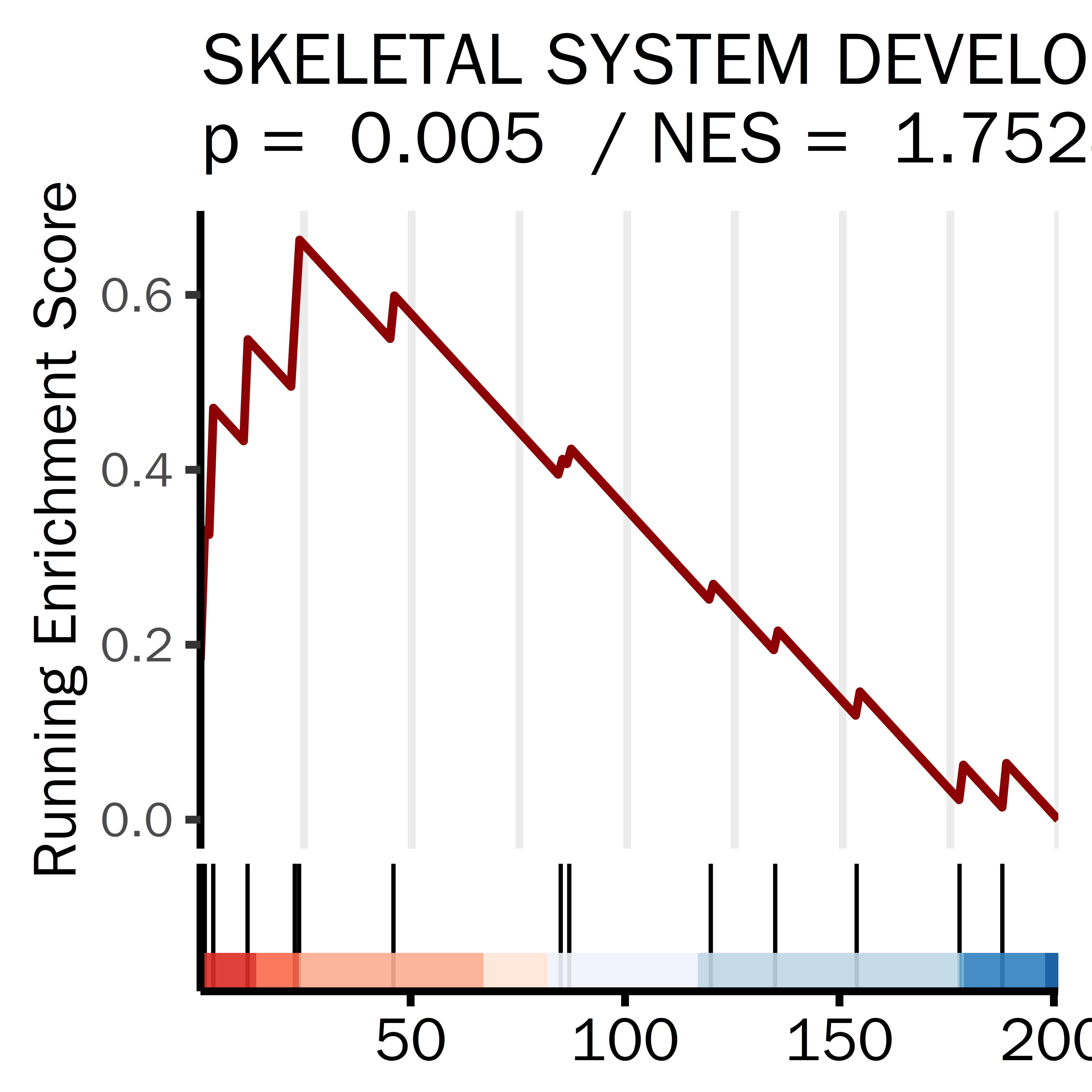
PLPLPLP = GSEA_result_pcut[grepl("GOBP_CELL_CYCLE",GSEA_result_pcut[,1]),]
datassssset = PLPLPLP
datassssset = datassssset[1,]
name1 = "CELL CYCLE"
i = 1
ID = datassssset[i,1]
gseaplot2(GSEA_result,
rel_heights = c(0.5, 0.1, 0.1),
geneSetID = ID,
base_size = 20,
color = "darkblue",
title =paste(name1,
"\np = ",
round(datassssset[i,5],4),
" / NES = ",
round(datassssset[i,4],4)),
subplots = 1:2,
ES_geom = "line")
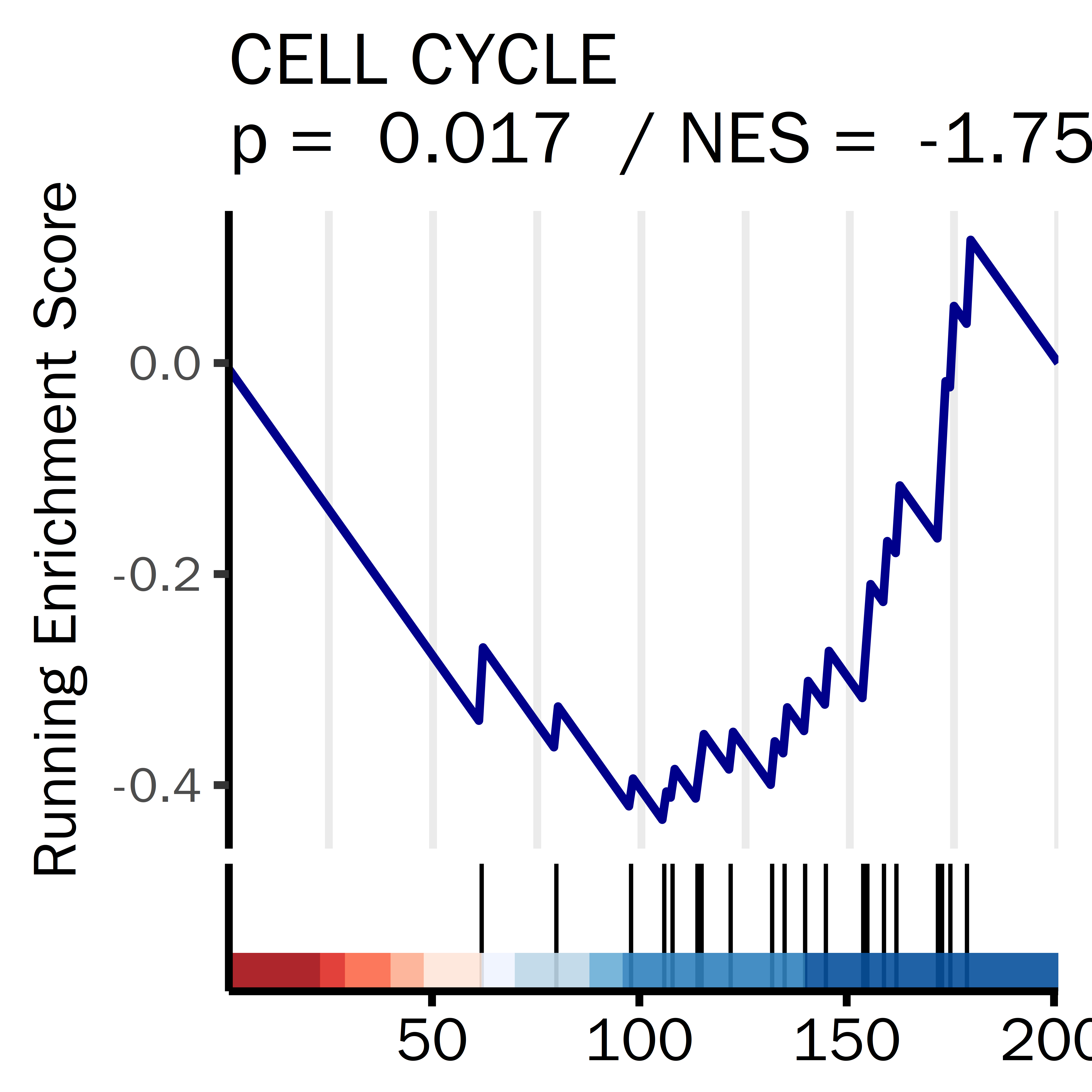
PLPLPLP = GSEA_result_pcut[grepl("GOBP_CELL_CELL_ADHESION",GSEA_result_pcut[,1]),]
datassssset = PLPLPLP
datassssset = datassssset[1,]
name1 = "CELL CELL ADHESION"
i = 1
ID = datassssset[i,1]
gseaplot2(GSEA_result,
rel_heights = c(0.5, 0.1, 0.1),
geneSetID = ID,
base_size = 20,
color = "darkblue",
title =paste(name1,
"\np = ",
round(datassssset[i,5],4),
" / NES = ",
round(datassssset[i,4],4)),
subplots = 1:2,
ES_geom = "line")