2F-G
Shift_100kb = read.table("/data3/psg/Ets1/ProcessedData/ShiftCOMP_100kbDATA.txt", sep = "\t", header=T,fill = TRUE)
ShiftTO_A_100kb = Shift_100kb[which(Shift_100kb$comp == "BtoA"),]
ShiftTO_B_100kb = Shift_100kb[which(Shift_100kb$comp == "AtoB"),]
ShiftTO_A_100kb_gene = ShiftTO_A_100kb$gene
ShiftTO_A_100kb_gene = as.data.frame(ShiftTO_A_100kb_gene)
colnames(ShiftTO_A_100kb_gene) = "gene"
ShiftTO_A_100kb_gene = unique(ShiftTO_A_100kb_gene)
ShiftTO_B_100kb_gene = ShiftTO_B_100kb$gene
ShiftTO_B_100kb_gene = as.data.frame(ShiftTO_B_100kb_gene)
colnames(ShiftTO_B_100kb_gene) = "gene"
ShiftTO_B_100kb_gene = unique(ShiftTO_B_100kb_gene)
Diff_0TO4_DEG_FULL = read.table("/Ets1/ProcessedData/Stage1_DEAdata.txt",sep = "\t", header=T,fill = TRUE)
Diff_0TO4_DEG_tmp = Diff_0TO4_DEG_FULL[which(Diff_0TO4_DEG_FULL[,7] <= 0.0001),]
Diff_0TO4_DEG_UP = Diff_0TO4_DEG_tmp[which(Diff_0TO4_DEG_tmp[,3] >= log2(1.5)),]
Diff_0TO4_DEG_DOWN = Diff_0TO4_DEG_tmp[which(Diff_0TO4_DEG_tmp[,3] <= -log2(1.5)),]
Diff_0TO4_SEG = Diff_0TO4_DEG_FULL[which(Diff_0TO4_DEG_FULL[,7] >= 0.2),]
Diff_0TO4_DEG_UP_gene = Diff_0TO4_DEG_UP[,1]
Diff_0TO4_DEG_UP_gene = as.data.frame(Diff_0TO4_DEG_UP_gene)
colnames(Diff_0TO4_DEG_UP_gene) = "gene"
ShiftTO_A_100kb_UPgene = inner_join(ShiftTO_A_100kb_gene,Diff_0TO4_DEG_UP_gene,by="gene")
ShiftTO_B_100kb_DOWNgene = inner_join(ShiftTO_B_100kb_gene,Diff_0TO4_DEG_DOWN,by="gene")
geneet = rbind(ShiftTO_A_100kb_UPgene)
ego <- groupGO(gene = geneet[,1],
OrgDb = org.Mm.eg.db,
ont = "BP",
level = 1,
readable = F)
level1GO = as.data.frame(ego)
level1GO = level1GO[,2]
level1GO = as.data.frame(level1GO)
colnames(level1GO) = "Description"
ego <- groupGO(gene = geneet[,1],
OrgDb = org.Mm.eg.db,
ont = "BP",
level = 2,
readable = F)
level2GO = as.data.frame(ego)
level2GO = level2GO[,2]
level2GO = as.data.frame(level2GO)
colnames(level2GO) = "Description"
ego <- groupGO(gene = geneet[,1],
OrgDb = org.Mm.eg.db,
ont = "BP",
level = 3,
readable = F)
level3GO = as.data.frame(ego)
level3GO = level3GO[,2]
level3GO = as.data.frame(level3GO)
colnames(level3GO) = "Description"
ego <- groupGO(gene = geneet[,1],
OrgDb = org.Mm.eg.db,
ont = "BP",
level = 4,
readable = F)
level4GO = as.data.frame(ego)
level4GO = level4GO[,2]
level4GO = as.data.frame(level4GO)
colnames(level4GO) = "Description"
ego <- groupGO(gene = geneet[,1],
OrgDb = org.Mm.eg.db,
ont = "BP",
level = 5,
readable = F)
level5GO = as.data.frame(ego)
level5GO = level5GO[,2]
level5GO = as.data.frame(level5GO)
colnames(level5GO) = "Description"
levelGO = rbind(level1GO,level2GO,level3GO,level4GO)
levelGO = unique(levelGO)
levelGO = as.data.frame(levelGO)
colnames(levelGO) = "Description"
ego <- enrichGO(gene = ShiftTO_A_100kb_UPgene[,1],
OrgDb = org.Mm.eg.db,
keyType = 'SYMBOL',
ont = "BP")
egoplotdata1 <- mutate(ego, richFactor = Count / as.numeric(sub("/\\d+", "", BgRatio)))
egoplotdata2 <- mutate(egoplotdata1, BgRatio_num = as.numeric(sub("/\\d+", "", BgRatio))/1)
GOdata <- mutate(egoplotdata2, GeneRatio_num = as.numeric(sub("/\\d+", "", GeneRatio))/1)
ShiftTO_A_100kb_UPgene_GO = as.data.frame(GOdata)
ShiftTO_A_100kb_UPgene_GO = inner_join(ShiftTO_A_100kb_UPgene_GO,levelGO,by="Description")
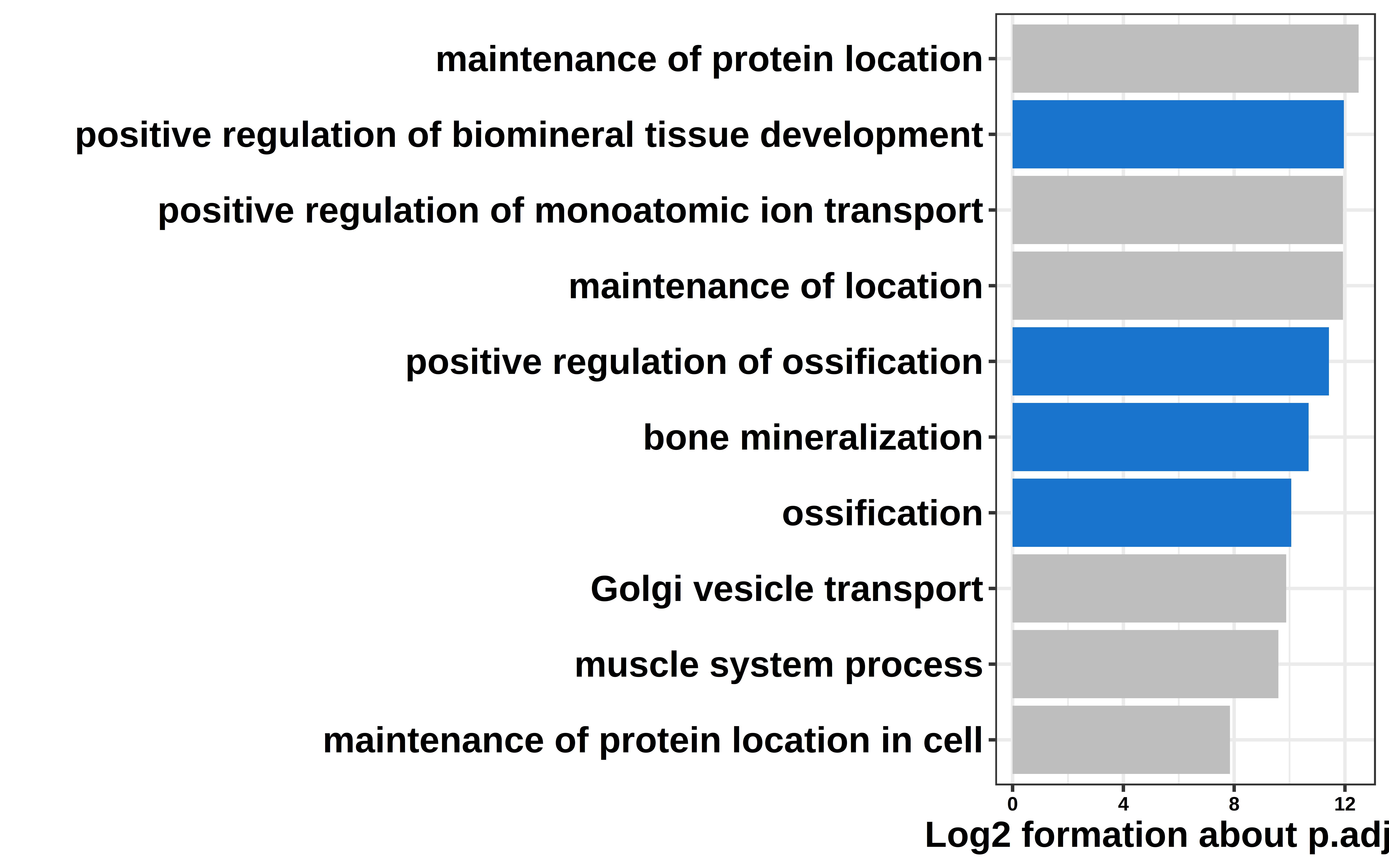
ShiftTO_A_100kb_UPgene_GO_top10 = ShiftTO_A_100kb_UPgene_GO[1:10,]
ego <- enrichGO(gene = ShiftTO_B_100kb_DOWNgene[,1],
OrgDb = org.Mm.eg.db,
keyType = 'SYMBOL',
ont = "BP")
egoplotdata1 <- mutate(ego, richFactor = Count / as.numeric(sub("/\\d+", "", BgRatio)))
egoplotdata2 <- mutate(egoplotdata1, BgRatio_num = as.numeric(sub("/\\d+", "", BgRatio))/1)
GOdata <- mutate(egoplotdata2, GeneRatio_num = as.numeric(sub("/\\d+", "", GeneRatio))/1)
ShiftTO_B_100kb_DOWNgene_GO = as.data.frame(GOdata)
ShiftTO_B_100kb_DOWNgene_GO = inner_join(ShiftTO_B_100kb_DOWNgene_GO,levelGO,by="Description")
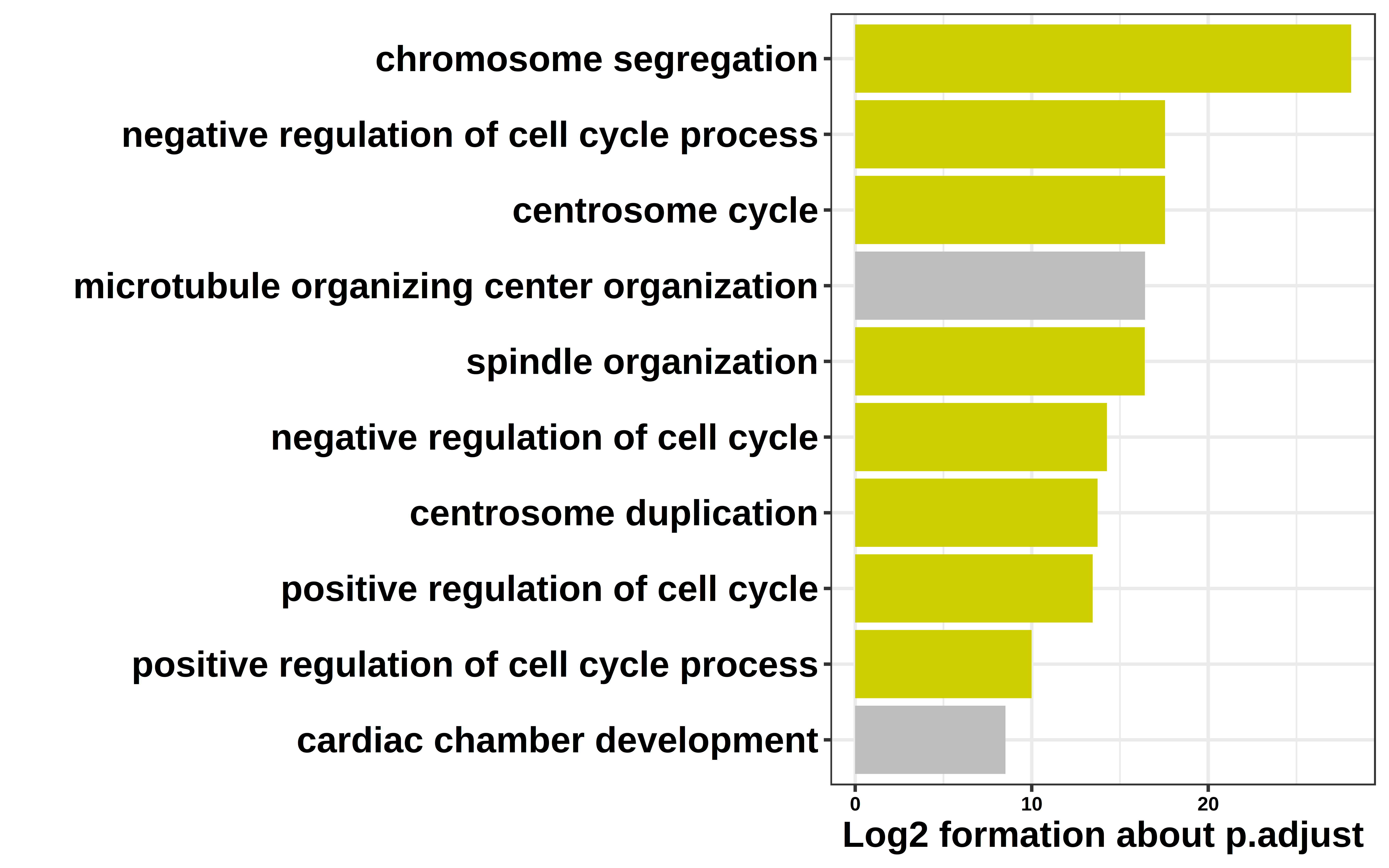
ShiftTO_B_100kb_DOWNgene_GO_top10 = ShiftTO_B_100kb_DOWNgene_GO[1:10,]
ShiftTO_B_100kb_DOWNgene_GO_top10$NP = log2(ShiftTO_B_100kb_DOWNgene_GO_top10[,6])*(-1)
col = c("CC","CC","CC","NO","CC","CC","CC","CC","CC","NO")
col = as.data.frame(col)
ShiftTO_B_100kb_DOWNgene_GO_top10 = cbind(ShiftTO_B_100kb_DOWNgene_GO_top10,col)
ShiftTO_A_100kb_UPgene_GO_top10$NP = log2(ShiftTO_A_100kb_UPgene_GO_top10[,6])*(-1)
col = c("NO","bone","NO","NO","bone","bone","bone","NO","NO","NO")
col = as.data.frame(col)
ShiftTO_A_100kb_UPgene_GO_top10 = cbind(ShiftTO_A_100kb_UPgene_GO_top10,col)
options(repr.plot.width = 8, repr.plot.height = 5, repr.plot.res = 1000, repr.plot.pointsize = 10)
ggplot(ShiftTO_A_100kb_UPgene_GO_top10,
aes(NP, fct_reorder(Description, NP),fill=col)) +
geom_col() +
scale_fill_manual(values = c("dodgerblue3","grey"))+
theme_dose(12) +
ylab("\n") +
xlab("Log2 formation about p.adjust") +
theme(plot.title=element_text(face="bold",hjust=0.5,size=25,color = "darkblue"),
axis.text.x=element_text(face="bold",size=8),
axis.text.y=element_text(face="bold",size=15),
axis.title.x = element_text(face="bold",size = 15),
legend.title=element_text(face="bold",size=10),
legend.position="none",
legend.text=element_text(face="bold",size=8))
ggplot(ShiftTO_B_100kb_DOWNgene_GO_top10,
aes(NP, fct_reorder(Description, NP),fill=col)) +
geom_col() +
scale_fill_manual(values = c("yellow3","grey"))+
theme_dose(12) +
ylab("\n") +
xlab("Log2 formation about p.adjust") +
theme(plot.title=element_text(face="bold",hjust=0.5,size=25,color = "darkblue"),
axis.text.x=element_text(face="bold",size=8),
axis.text.y=element_text(face="bold",size=15),
axis.title.x = element_text(face="bold",size = 15),
legend.title=element_text(face="bold",size=10),
legend.position="none",
legend.text=element_text(face="bold",size=8))
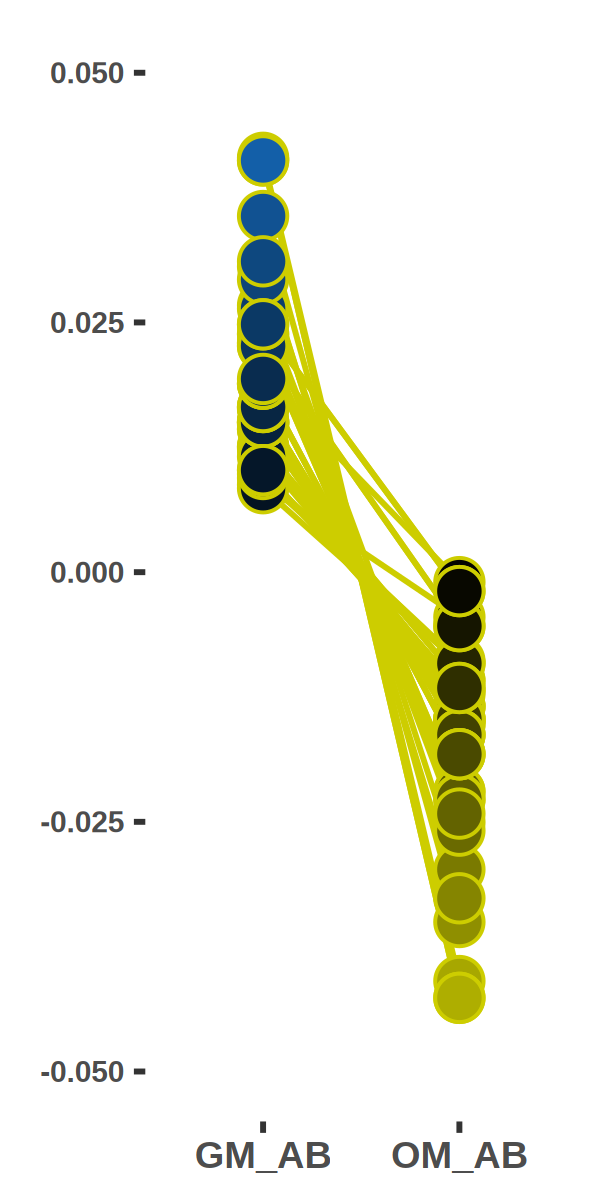
ShiftTO_B_100kb_DOWNgene_GO_top10_line = ShiftTO_B_100kb_DOWNgene_GO_top10[which(ShiftTO_B_100kb_DOWNgene_GO_top10$col == "CC"),]
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene = ShiftTO_B_100kb_DOWNgene_GO_top10_line$geneID
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene = as.data.frame(ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene)
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene_vec = paste(ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[1,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[2,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[3,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[4,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[5,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[6,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[7,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[8,1],
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene[9,1],
sep="/")
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene = as.data.frame(strsplit(ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene_vec, split = "/"))
colnames(ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene) = "gene"
ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene = unique(ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene)
ShiftTO_B_100kb = inner_join(ShiftTO_B_100kb,ShiftTO_B_100kb_DOWNgene_GO_top10_line_gene,by="gene")
ShiftTO_B_100kb = ShiftTO_B_100kb[,c(8:18,30)]
ShiftTO_B_100kb$ID = c(1:nrow(ShiftTO_B_100kb))
f1 = as.data.frame(cbind(ShiftTO_B_100kb[1,13],"GM_AB",ShiftTO_B_100kb[1,4]))
f2 = as.data.frame(cbind(ShiftTO_B_100kb[1,13],"OM_AB",ShiftTO_B_100kb[1,5]))
plot = rbind(f1,f2)
colnames(plot) = c("ID","condition","value")
plot[,3] = as.numeric(plot[,3])
for(i in 2:nrow(ShiftTO_B_100kb)){
f1 = as.data.frame(cbind(ShiftTO_B_100kb[i,13],"GM_AB",ShiftTO_B_100kb[i,4]))
f2 = as.data.frame(cbind(ShiftTO_B_100kb[i,13],"OM_AB",ShiftTO_B_100kb[i,5]))
plot_frag = rbind(f1,f2)
colnames(plot_frag) = c("ID","condition","value")
plot = rbind(plot,plot_frag)
}
plot[,3] = as.numeric(plot[,3])
ShiftTO_B_100kb$DIFF = ShiftTO_B_100kb[,5]-ShiftTO_B_100kb[,4]
ShiftTO_B_100kb = ShiftTO_B_100kb[order(ShiftTO_B_100kb[,14]),]
plot_ONE = plot[which(plot$ID == 14),]
options(repr.plot.width = 2, repr.plot.height = 4, repr.plot.res = 300, repr.plot.pointsize = 40)
ggplot(plot,aes(x=factor(condition), y=value, group=ID,fill=value)) +
geom_line(size=0.5,color = "yellow3") +
geom_point(shape=21,size=5,color = "yellow3") +
scale_fill_gradientn(colours=colorRampPalette(c("yellow3", "black", "dodgerblue3"))(100),limits = c(-0.05, 0.05))+
ylim(-0.05,0.05)+
xlab(NULL) +
ggtitle(NULL)+
ylab("") +
theme(plot.title=element_text(face="bold",hjust=0.5,size=25,color = "darkblue"),
axis.text.x=element_text(face="bold",size=9),
axis.text.y=element_text(face="bold",size=7),
axis.title.x = element_text(face="bold",size = 13),
axis.title.y = element_text(face="bold",size = 0,color = "darkblue"),
legend.title=element_text(face="bold",size=15),
legend.position="none",
legend.text=element_text(face="bold",size=15),
panel.background = element_rect(fill=NA, color=NA),
plot.background = element_rect(fill=NA, color=NA))
ggsave("/data3/psg/NGS_2025/4DN/HiC_data/DOWN_LINEALL.png",width = 2, height = 4)
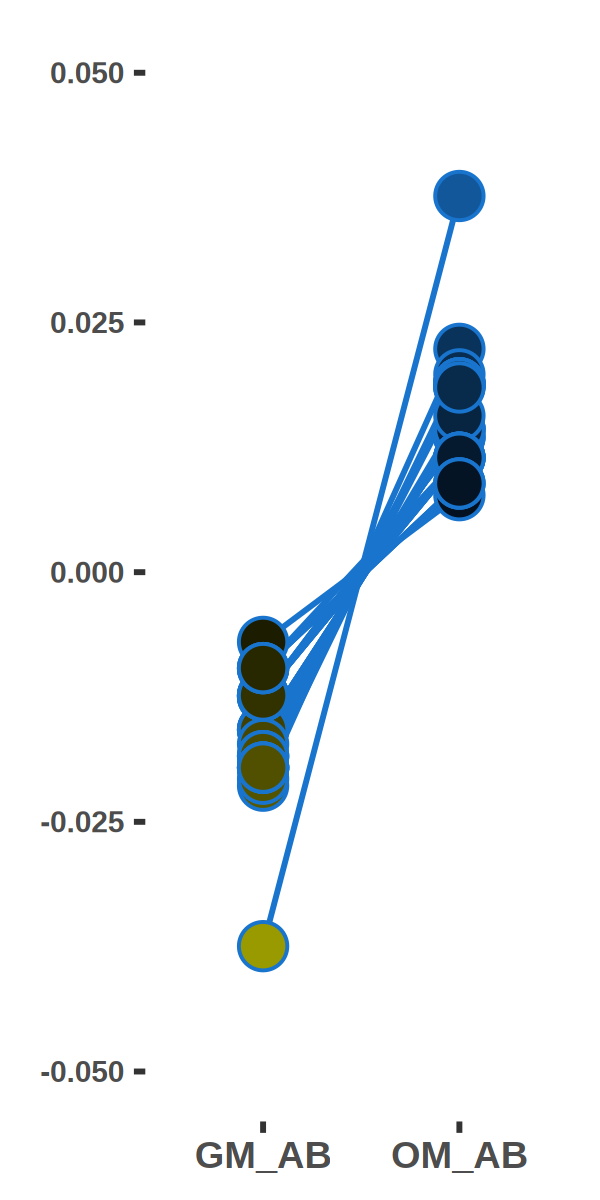
ShiftTO_A_100kb_UPgene_GO_top10_line = ShiftTO_A_100kb_UPgene_GO_top10[which(ShiftTO_A_100kb_UPgene_GO_top10$col == "bone"),]
ShiftTO_A_100kb_UPgene_GO_top10_line_gene = ShiftTO_A_100kb_UPgene_GO_top10_line$geneID
ShiftTO_A_100kb_UPgene_GO_top10_line_gene = as.data.frame(ShiftTO_A_100kb_UPgene_GO_top10_line_gene)
ShiftTO_A_100kb_UPgene_GO_top10_line_gene_vec = paste(ShiftTO_A_100kb_UPgene_GO_top10_line_gene[1,1],
ShiftTO_A_100kb_UPgene_GO_top10_line_gene[2,1],
ShiftTO_A_100kb_UPgene_GO_top10_line_gene[3,1],
ShiftTO_A_100kb_UPgene_GO_top10_line_gene[4,1],
sep="/")
ShiftTO_A_100kb_UPgene_GO_top10_line_gene = as.data.frame(strsplit(ShiftTO_A_100kb_UPgene_GO_top10_line_gene_vec, split = "/"))
colnames(ShiftTO_A_100kb_UPgene_GO_top10_line_gene) = "gene"
ShiftTO_A_100kb_UPgene_GO_top10_line_gene = unique(ShiftTO_A_100kb_UPgene_GO_top10_line_gene)
ShiftTO_A_100kb = inner_join(ShiftTO_A_100kb,ShiftTO_A_100kb_UPgene_GO_top10_line_gene,by="gene")
ShiftTO_A_100kb = ShiftTO_A_100kb[,c(8:18,30)]
ShiftTO_A_100kb$ID = c(1:nrow(ShiftTO_A_100kb))
f1 = as.data.frame(cbind(ShiftTO_A_100kb[1,13],"GM_AB",ShiftTO_A_100kb[1,4]))
f2 = as.data.frame(cbind(ShiftTO_A_100kb[1,13],"OM_AB",ShiftTO_A_100kb[1,5]))
plot = rbind(f1,f2)
colnames(plot) = c("ID","condition","value")
plot[,3] = as.numeric(plot[,3])
for(i in 2:nrow(ShiftTO_A_100kb)){
f1 = as.data.frame(cbind(ShiftTO_A_100kb[i,13],"GM_AB",ShiftTO_A_100kb[i,4]))
f2 = as.data.frame(cbind(ShiftTO_A_100kb[i,13],"OM_AB",ShiftTO_A_100kb[i,5]))
plot_frag = rbind(f1,f2)
colnames(plot_frag) = c("ID","condition","value")
plot = rbind(plot,plot_frag)
}
plot[,3] = as.numeric(plot[,3])
ShiftTO_A_100kb$DIFF = ShiftTO_A_100kb[,5]-ShiftTO_A_100kb[,4]
ShiftTO_A_100kb = ShiftTO_A_100kb[order(-ShiftTO_A_100kb[,14]),]
plot_ONE = plot[which(plot$ID == 6),]
options(repr.plot.width = 2, repr.plot.height = 4, repr.plot.res = 300, repr.plot.pointsize = 40)
ggplot(plot,aes(x=factor(condition), y=value, group=ID,fill=value)) +
geom_line(size=0.5,color = "dodgerblue3") +
geom_point(shape=21,size=5,color = "dodgerblue3") +
scale_fill_gradientn(colours=colorRampPalette(c("yellow3", "black", "dodgerblue3"))(100),limits = c(-0.05, 0.05))+
#theme_classic(base_size = 10) +
ylim(-0.05,0.05)+
xlab(NULL) +
ggtitle(NULL)+
ylab("") +
theme(plot.title=element_text(face="bold",hjust=0.5,size=25,color = "darkblue"),
axis.text.x=element_text(face="bold",size=9),
axis.text.y=element_text(face="bold",size=7),
axis.title.x = element_text(face="bold",size = 13),
axis.title.y = element_text(face="bold",size = 0,color = "darkblue"),
legend.title=element_text(face="bold",size=15),
legend.position="none",
legend.text=element_text(face="bold",size=15),
panel.background = element_rect(fill=NA, color=NA),
plot.background = element_rect(fill=NA, color=NA))
ggsave("/data3/psg/NGS_2025/4DN/HiC_data/DOWN_LINEALL.png",width = 2, height = 4)