3B-C
Stage1_UP_GO = read.table("/Ets1/ProcessedData/Stage1_UP_GO.txt", sep = "\t", header=T,fill = TRUE)
GENE_to_EpiSignal = read.table("/Ets1/ProcessedData/GENE_to_EpiSignal.txt", sep = "\t", header=T,fill = TRUE)
reg_gene_GO_bone = rbind(Stage1_UP_GO[grepl("ossification",Stage1_UP_GO[,2]), ],
Stage1_UP_GO[grepl("osteoblast differentiation",Stage1_UP_GO[,2]), ],
Stage1_UP_GO[grepl("Wnt",Stage1_UP_GO[,2]), ],
Stage1_UP_GO[grepl("bone",Stage1_UP_GO[,2]), ])
reg_gene_GO_bone = reg_gene_GO_bone[!grepl("regulation",reg_gene_GO_bone[,2]), ]
reg_gene_GO_bone = reg_gene_GO_bone[!grepl("propliferation",reg_gene_GO_bone[,2]), ]
reg_gene_GO_bone = unique(reg_gene_GO_bone)
reg_gene_GO_bone = reg_gene_GO_bone[order(-reg_gene_GO_bone$richFactor),]
reg_gene_GO_bone_gene = as.data.frame(strsplit(reg_gene_GO_bone[1,11], split = '/'))
colnames(reg_gene_GO_bone_gene) = "gene"
reg_gene_GO_bone_gene = unique(reg_gene_GO_bone_gene)
for(i in 1:nrow(reg_gene_GO_bone)){
reg_gene_GO_bone_gene_frag = as.data.frame(strsplit(reg_gene_GO_bone[i,11], split = '/'))
colnames(reg_gene_GO_bone_gene_frag) = "gene"
reg_gene_GO_bone_gene_frag = unique(reg_gene_GO_bone_gene_frag)
reg_gene_GO_bone_gene = rbind(reg_gene_GO_bone_gene,reg_gene_GO_bone_gene_frag)
}
reg_gene_GO_bone_gene = unique(reg_gene_GO_bone_gene)
reg_gene_GO_bone_gene_to_signal = inner_join(GENE_to_EpiSignal,reg_gene_GO_bone_gene,by="gene")
reg_gene_GO_bone_gene_to_signal = na.omit(reg_gene_GO_bone_gene_to_signal)
reg_gene_GO_bone_gene_to_signal = reg_gene_GO_bone_gene_to_signal[,c(1:4,12:19,23:29,31)]
options(repr.plot.width = 2, repr.plot.height = 3, repr.plot.res = 1000, repr.plot.pointsize = 20)
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardA"),c(1,14)]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardB"),c(1,14)]
reg_gene_GO_bone_gene_to_signal_toA[,1] = "toward\nA"
reg_gene_GO_bone_gene_to_signal_toB[,1] = "toward\nB"
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal_toA[order(-reg_gene_GO_bone_gene_to_signal_toA[,2]),]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal_toB[order(-reg_gene_GO_bone_gene_to_signal_toB[,2]),]
violin_plot = rbind(reg_gene_GO_bone_gene_to_signal_toA,reg_gene_GO_bone_gene_to_signal_toB)
colnames(violin_plot) = c("comp","value")
violin_plot$norm = (violin_plot[,2] - mean(violin_plot[,2]))/(sd(violin_plot[,2]))
violin_plot = violin_plot[which(violin_plot$norm >= -3),]
violin_plot = violin_plot[which(violin_plot$norm <= 3),]
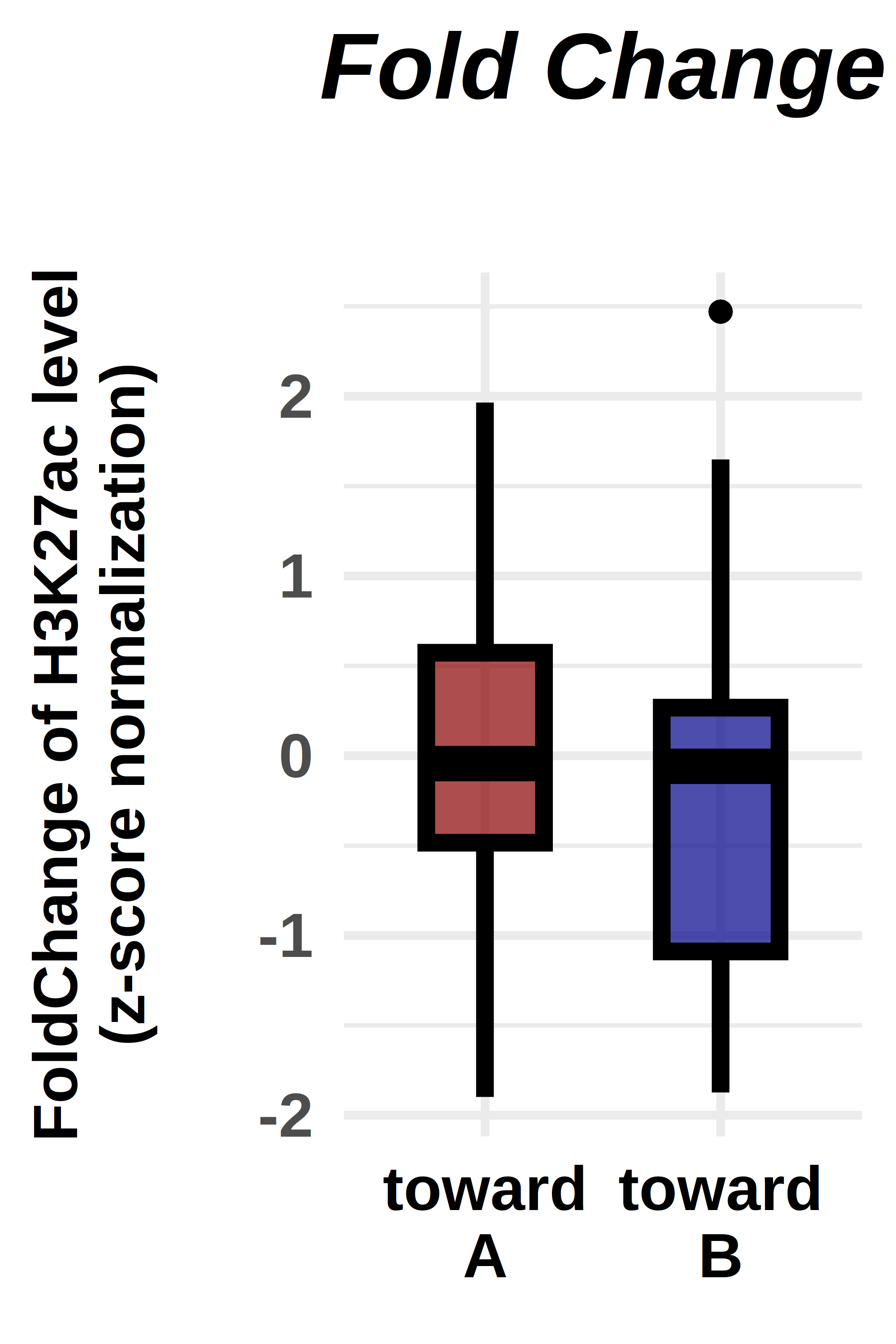
ggplot(violin_plot, aes(x=factor(comp), y=norm, fill=factor(comp)))+
geom_boxplot(fill=c("darkred","darkblue"),
size=1,
color='black',
width=0.5,
alpha=0.7)+
guides(fill="none")+
theme_minimal()+
labs(title="Fold Change",
subtitle = "")+
xlab("")+
ylab("FoldChange of H3K27ac level\n(z-score normalization)\n")+
theme(plot.title=element_text(face="bold.italic",hjust=0.5,size=15,color = "black"),
axis.text.x=element_text(face="bold",size=10,color = "black"),
axis.text.y=element_text(face="bold",size=10),
plot.subtitle=element_text(face="bold",hjust=0.5,vjust=-3,size=15,color = "black"),
axis.title.x = element_text(size=0,color = "black"),
axis.title.y = element_text(face="bold",size=10,color = "black"))
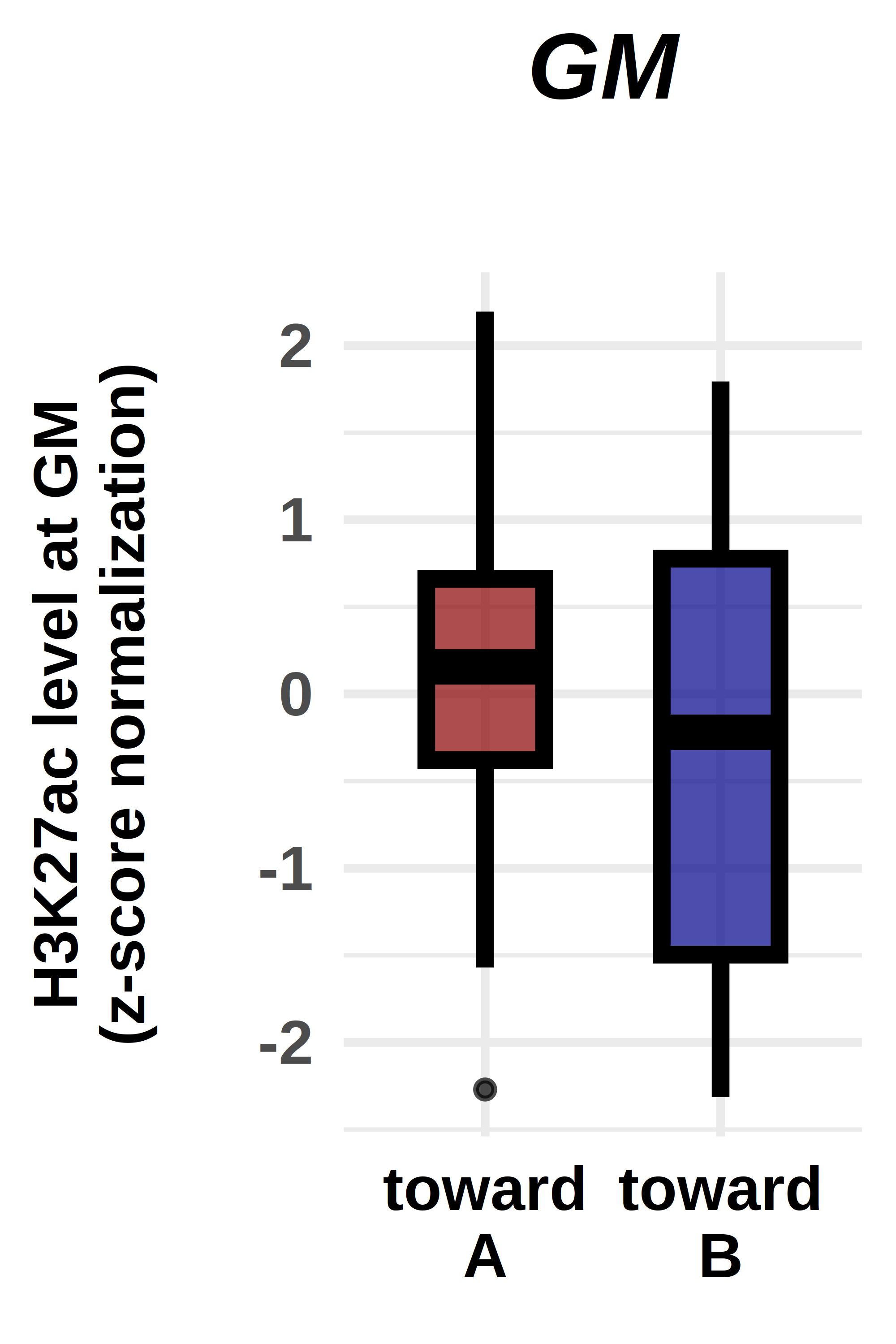
options(repr.plot.width = 2, repr.plot.height = 3, repr.plot.res = 1000, repr.plot.pointsize = 20)
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardA"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardB"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_toA[,1] = "toward\nA"
reg_gene_GO_bone_gene_to_signal_toB[,1] = "toward\nB"
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal_toA[order(-reg_gene_GO_bone_gene_to_signal_toA[,2]),]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal_toB[order(-reg_gene_GO_bone_gene_to_signal_toB[,2]),]
violin_plot = rbind(reg_gene_GO_bone_gene_to_signal_toA,reg_gene_GO_bone_gene_to_signal_toB)
colnames(violin_plot) = c("comp","value")
violin_plot$norm = (violin_plot[,2] - mean(violin_plot[,2]))/(sd(violin_plot[,2]))
violin_plot = violin_plot[which(violin_plot$norm >= -3),]
violin_plot = violin_plot[which(violin_plot$norm <= 3),]
ggplot(violin_plot, aes(x=factor(comp), y=norm, fill=factor(comp)))+
geom_boxplot(fill=c("darkred","darkblue"),
size=1,
color='black',
width=0.5,
alpha=0.7)+
guides(fill="none")+
theme_minimal()+
labs(title="GM",
subtitle = "")+
xlab("")+
ylab("H3K27ac level at GM\n(z-score normalization)\n")+
theme(plot.title=element_text(face="bold.italic",hjust=0.5,size=15,color = "black"),
axis.text.x=element_text(face="bold",size=10,color = "black"),
axis.text.y=element_text(face="bold",size=10),
plot.subtitle=element_text(face="bold",hjust=0.5,vjust=-3,size=15,color = "black"),
axis.title.x = element_text(size=0,color = "black"),
axis.title.y = element_text(face="bold",size=10,color = "black"))
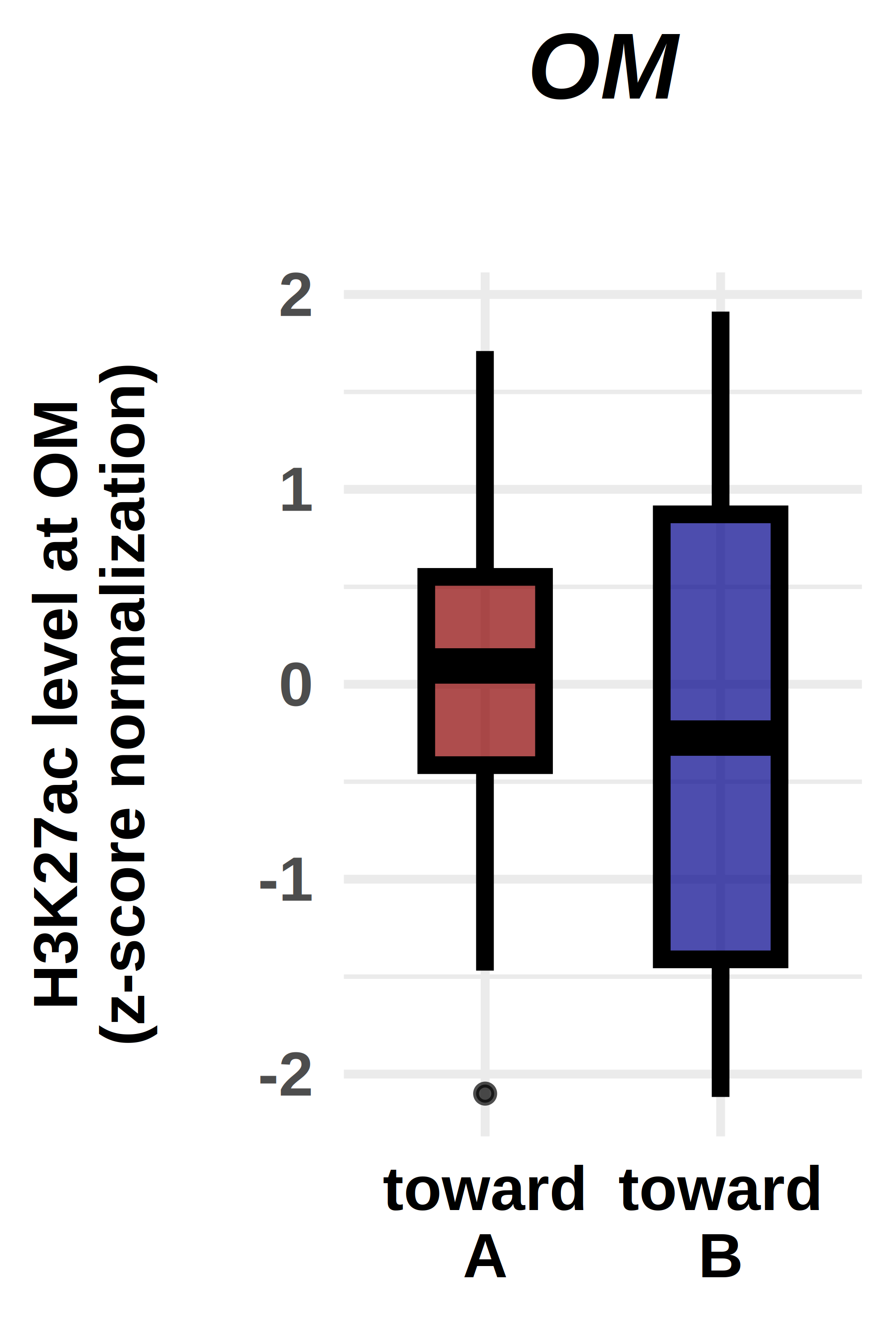
options(repr.plot.width = 2, repr.plot.height = 3, repr.plot.res = 1000, repr.plot.pointsize = 20)
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardA"),c(1,7)]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal$ABStatus == "towardB"),c(1,7)]
reg_gene_GO_bone_gene_to_signal_toA[,1] = "toward\nA"
reg_gene_GO_bone_gene_to_signal_toB[,1] = "toward\nB"
reg_gene_GO_bone_gene_to_signal_toA = reg_gene_GO_bone_gene_to_signal_toA[order(-reg_gene_GO_bone_gene_to_signal_toA[,2]),]
reg_gene_GO_bone_gene_to_signal_toB = reg_gene_GO_bone_gene_to_signal_toB[order(-reg_gene_GO_bone_gene_to_signal_toB[,2]),]
violin_plot = rbind(reg_gene_GO_bone_gene_to_signal_toA,reg_gene_GO_bone_gene_to_signal_toB)
colnames(violin_plot) = c("comp","value")
violin_plot$norm = (violin_plot[,2] - mean(violin_plot[,2]))/(sd(violin_plot[,2]))
violin_plot = violin_plot[which(violin_plot$norm >= -3),]
violin_plot = violin_plot[which(violin_plot$norm <= 3),]
ggplot(violin_plot, aes(x=factor(comp), y=norm, fill=factor(comp)))+
geom_boxplot(fill=c("darkred","darkblue"),
size=1,
color='black',
width=0.5,
alpha=0.7)+
guides(fill="none")+
theme_minimal()+
labs(title="OM",
subtitle = "")+
xlab("")+
ylab("H3K27ac level at OM\n(z-score normalization)\n")+
theme(plot.title=element_text(face="bold.italic",hjust=0.5,size=15,color = "black"),
axis.text.x=element_text(face="bold",size=10,color = "black"),
axis.text.y=element_text(face="bold",size=10),
plot.subtitle=element_text(face="bold",hjust=0.5,vjust=-3,size=15,color = "black"),
axis.title.x = element_text(size=0,color = "black"),
axis.title.y = element_text(face="bold",size=10,color = "black"))
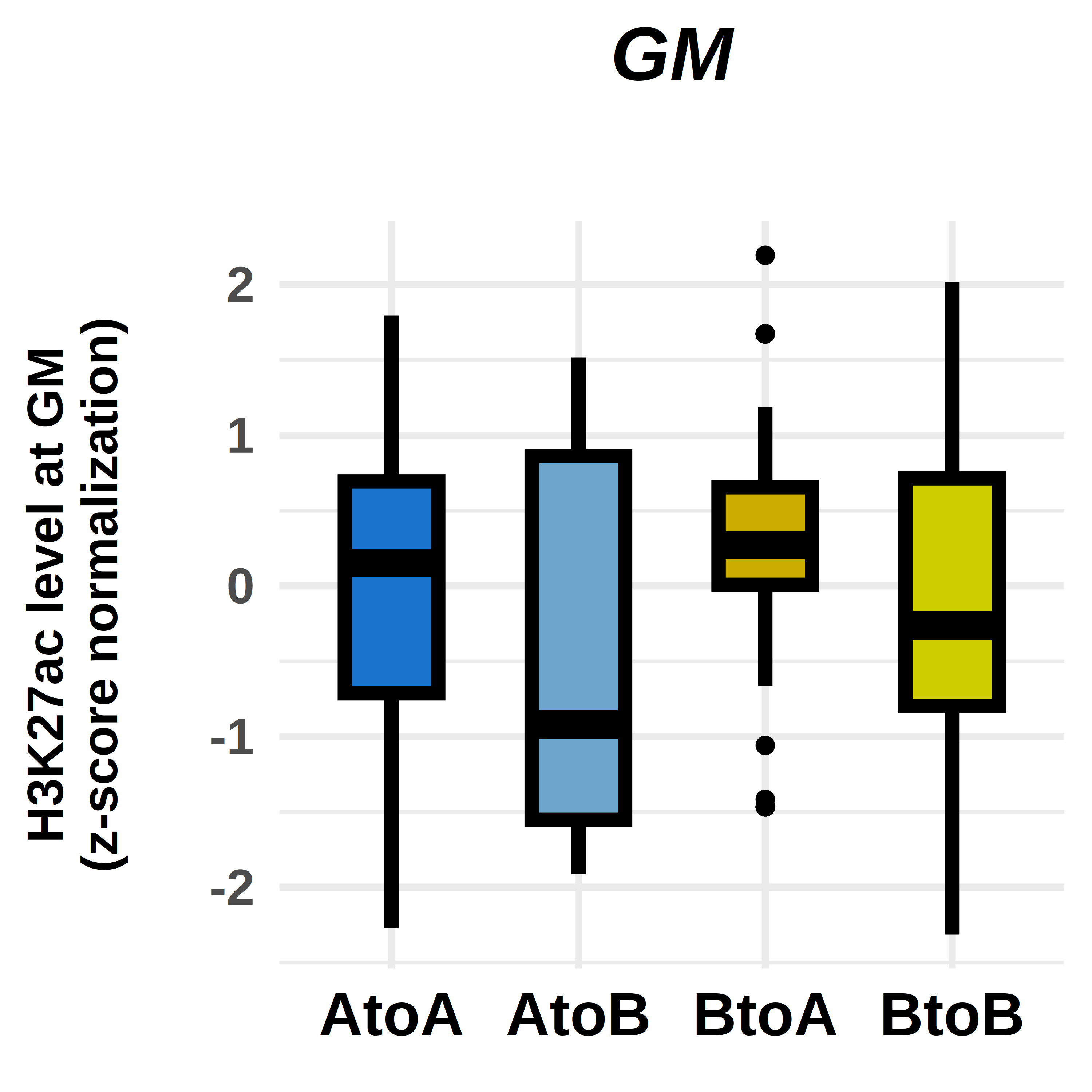
options(repr.plot.width = 3, repr.plot.height = 3, repr.plot.res = 1000, repr.plot.pointsize = 20)
reg_gene_GO_bone_gene_to_signal_AtoA = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal[,12] == "AtoA"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_BtoA = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal[,12] == "BtoA"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_AtoB = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal[,12] == "AtoB"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_BtoB = reg_gene_GO_bone_gene_to_signal[which(reg_gene_GO_bone_gene_to_signal[,12] == "BtoB"),c(1,6)]
reg_gene_GO_bone_gene_to_signal_AtoA[,1] = "AtoA"
reg_gene_GO_bone_gene_to_signal_BtoA[,1] = "BtoA"
reg_gene_GO_bone_gene_to_signal_AtoB[,1] = "AtoB"
reg_gene_GO_bone_gene_to_signal_BtoB[,1] = "BtoB"
violin_plot = rbind(reg_gene_GO_bone_gene_to_signal_AtoA,
reg_gene_GO_bone_gene_to_signal_BtoA,
reg_gene_GO_bone_gene_to_signal_AtoB,
reg_gene_GO_bone_gene_to_signal_BtoB)
colnames(violin_plot) = c("comp","value")
violin_plot$norm = (violin_plot[,2] - mean(violin_plot[,2]))/(sd(violin_plot[,2]))
violin_plot = violin_plot[which(violin_plot$norm >= -3),]
violin_plot = violin_plot[which(violin_plot$norm <= 3),]
ggplot(violin_plot, aes(x=factor(comp), y=norm, fill=factor(comp)))+
geom_boxplot(fill=c("dodgerblue3","skyblue3","gold3","yellow3"),
size=1,
color='black',
width=0.5,
alpha=1)+
guides(fill="none")+
theme_minimal()+
labs(title="GM",
subtitle = "")+
xlab("")+
ylab("H3K27ac level at GM\n(z-score normalization)\n")+
#ylim(0,3)+
theme(plot.title=element_text(face="bold.italic",hjust=0.5,size=15,color = "black"),
axis.text.x=element_text(face="bold",size=12,color = "black"),
axis.text.y=element_text(face="bold",size=10),
plot.subtitle=element_text(face="bold",hjust=0.5,vjust=-3,size=15,color = "black"),
axis.title.x = element_text(size=0,color = "black"),
axis.title.y = element_text(face="bold",size=10,color = "black"))