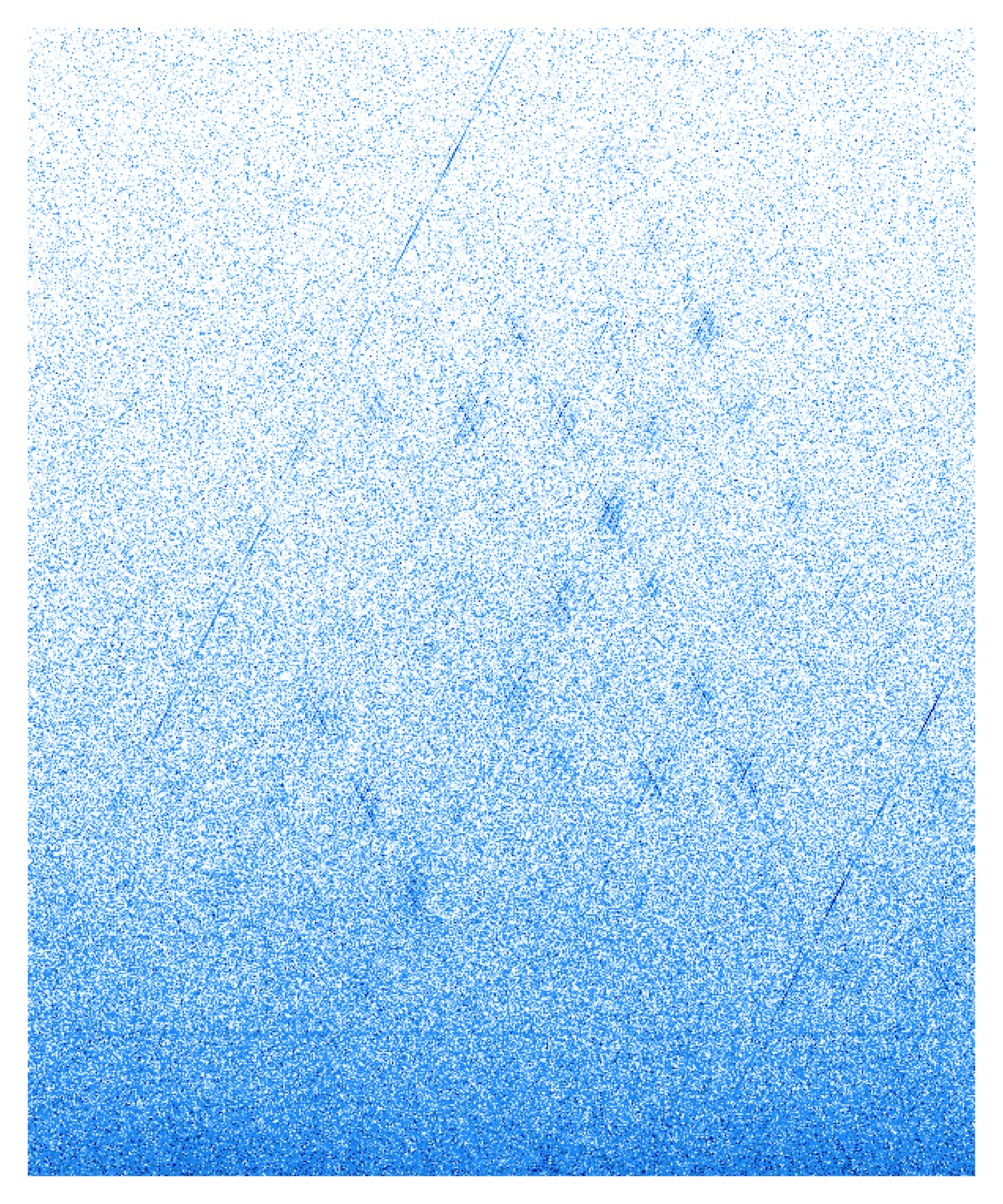
single_Vplot = function(input){
v <- read_vplot(input)
v$label = c(nrow(v):1)
v = v[order(v$label),-ncol(v)]
data = v
options(repr.plot.width = 2.5, repr.plot.height = 3, repr.plot.res = 1500, repr.plot.pointsize = 10)
conte=data
namen = c(1:nrow(data))
rownames(conte)=as.vector(namen)
PCA=conte
vvv=pheatmap(PCA,
cluster_rows = F,
cluster_cols = F,
show_rownames = F,
show_colnames = F,
legend = FALSE,
border_color = FALSE,
color = colorRampPalette(c("white","dodgerblue1","dodgerblue2","dodgerblue3","dodgerblue4","darkblue"))(100),
breaks=seq(from=0,to=2,length.out = 100))
vvv
}
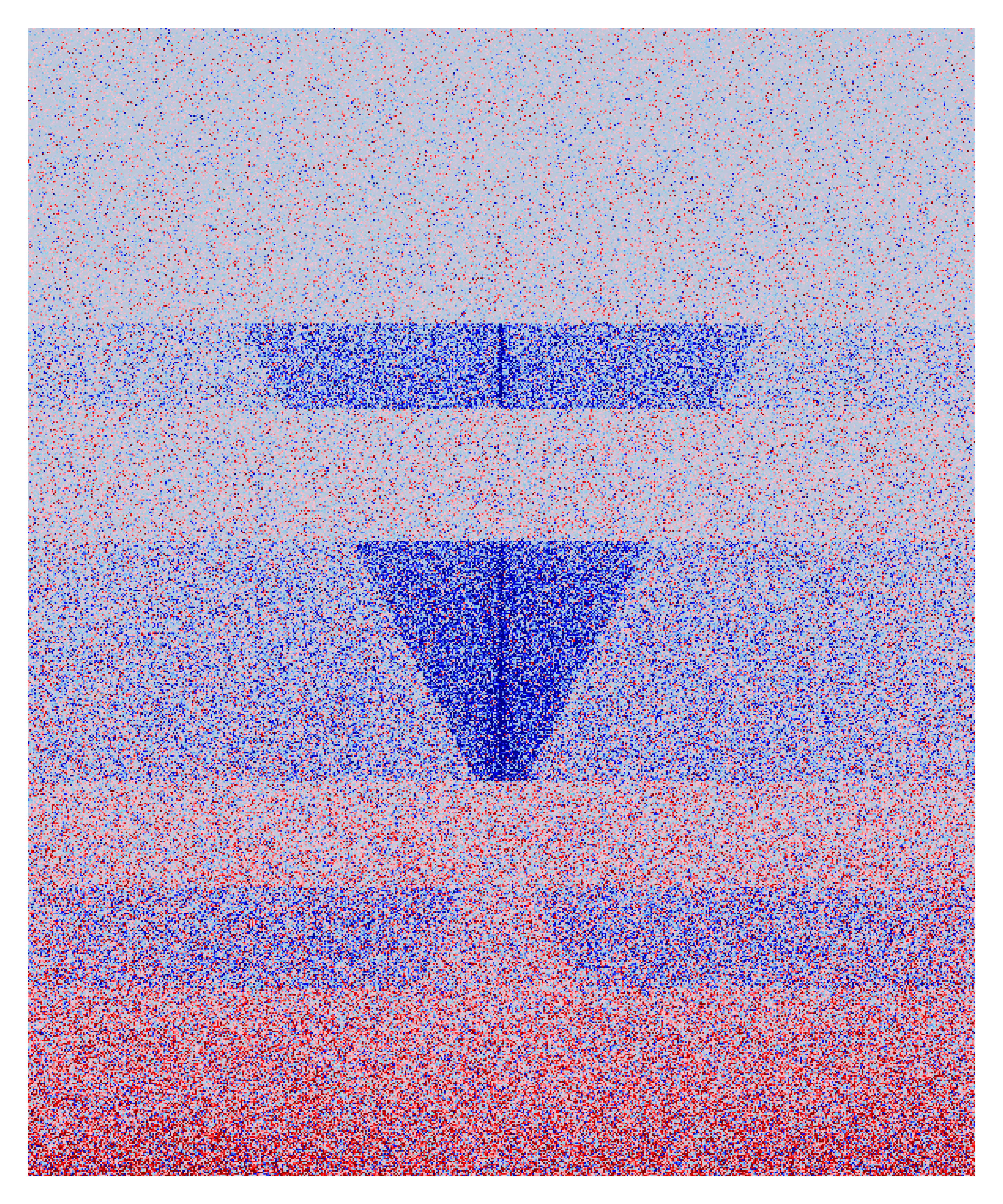
diff_Vplot = function(DAY0,DAY4){
DAY0 <- read_vplot(DAY0)
DAY4 <- read_vplot(DAY4)
v = DAY4 - DAY0
v$label = c(nrow(v):1)
v = v[order(v$label),-ncol(v)]
data = v
options(repr.plot.width = 2.5, repr.plot.height = 3, repr.plot.res = 1500, repr.plot.pointsize = 10)
conte=data
namen = c(1:nrow(data))
rownames(conte)=as.vector(namen)
PCA=conte
vvv=pheatmap(PCA,
cluster_rows = F,
cluster_cols = F,
show_rownames = F,
show_colnames = F,
legend = FALSE,
border_color = FALSE,
color = colorRampPalette(c("darkblue","blue","skyblue","pink","red","darkred"))(100),
breaks=seq(from=-1,to=1,length.out = 100))
vvv
}
# LINUX cmd
# pyatac vplot \
# --bam /data3/psg/Ets1/DAY0_ATAC.bam \
# --bed /data3/psg/Ets1/DAY0_NOR_Ets1.txt \
# --out /data3/psg/Ets1/DAY0_NOR_Ets1_50_800_300_10_5_V --cores 7 --lower 50 --upper 800 --flank 300 --weight 10 --strand 5 --scale
# pyatac vplot \
# --bam /data3/psg/Ets1/DAY4_ATAC.bam \
# --bed /data3/psg/Ets1/DAY4_NOR_Ets1.txt \
# --out /data3/psg/Ets1/DAY4_NOR_Ets1_50_800_300_10_5_V --cores 7 --lower 50 --upper 800 --flank 300 --weight 10 --strand 5 --scale
single_Vplot(input="/data3/psg/Ets1/DAY0_NOR_Ets1_50_800_300_10_5_V.Mat")
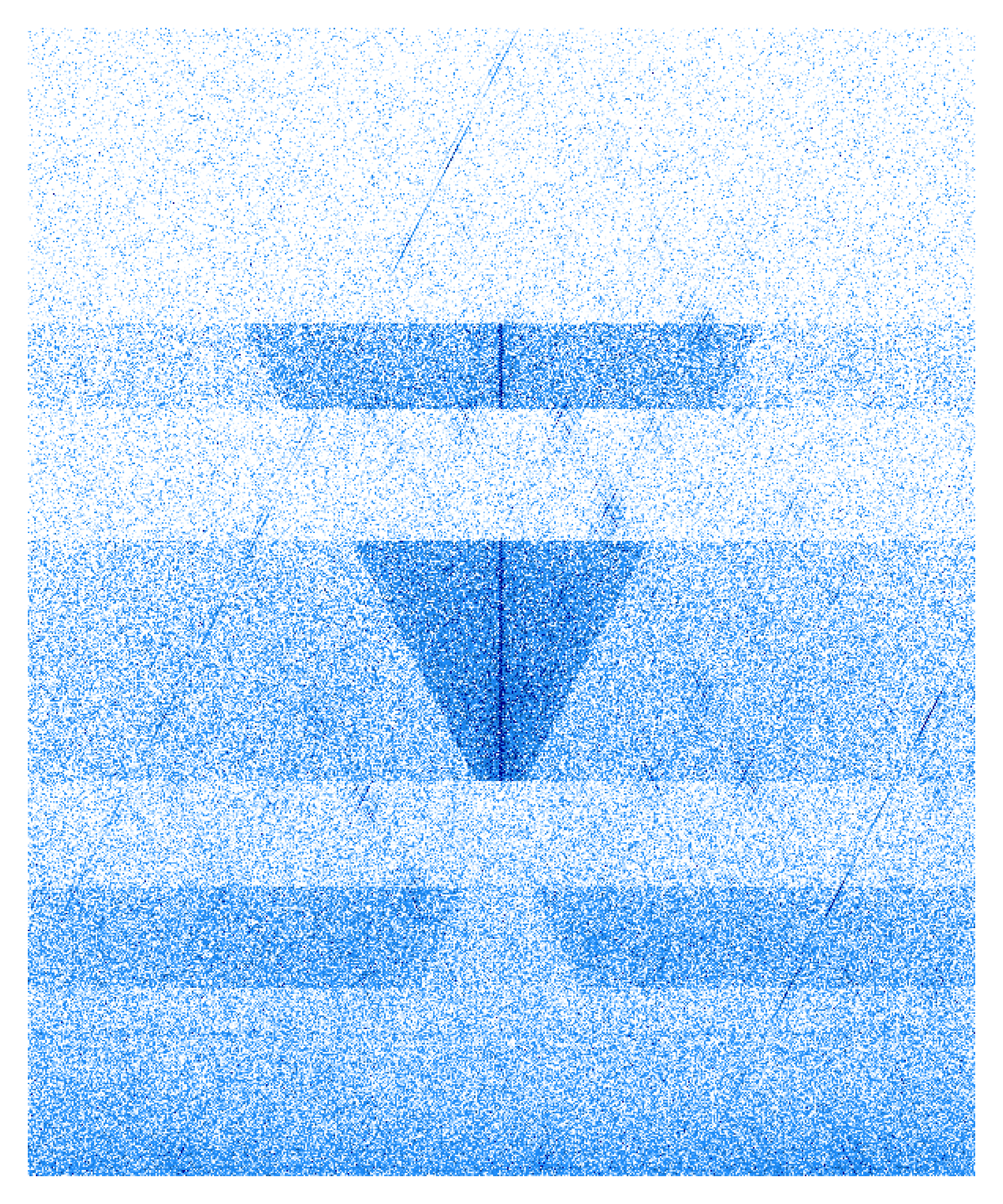
single_Vplot(input="/data3/psg/Ets1/DAY4_NOR_Ets1_50_800_300_10_5_V.Mat")
diff_Vplot(DAY0 = "/data3/psg/Ets1/DAY0_NOR_Ets1_50_800_300_10_5_V.Mat",
DAY4 = "/data3/psg/Ets1/DAY4_NOR_Ets1_50_800_300_10_5_V.Mat")