UMAP_DIFF_correlation_50_2_1_1 = as.data.frame(vroom("/Ets1/ProcessedData/UMAP_DIFF_correlation_50_2_1_1.txt", delim = "\t",show_col_types = FALSE))
UMAP_DIFF_correlation_50_2_1_1_fit = UMAP_DIFF_correlation_50_2_1_1[,1:2]
UMAP_DIFF_correlation_50_2_1_1_label = paste(UMAP_DIFF_correlation_50_2_1_1[,3],UMAP_DIFF_correlation_50_2_1_1[,4],UMAP_DIFF_correlation_50_2_1_1[,5])
UMAP_DIFF_correlation_50_2_1_1_label = as.data.frame(UMAP_DIFF_correlation_50_2_1_1_label)
colnames(UMAP_DIFF_correlation_50_2_1_1_label) = "label"
result <- kmeans(UMAP_DIFF_correlation_50_2_1_1_fit, centers = 22, iter.max = 10000, algorithm = "MacQueen")
UMAP_DIFF_correlation_50_2_1_1 = cbind(UMAP_DIFF_correlation_50_2_1_1,as.data.frame(result$cluster))
colnames(UMAP_DIFF_correlation_50_2_1_1)[ncol(UMAP_DIFF_correlation_50_2_1_1)] = "cluster"
num_cluster = 1
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
AB_DIFF = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
AB_DIFF = as.data.frame(AB_DIFF)
colnames(AB_DIFF) = "AB_DIFF"
AB_DIFF$score = 0
for(i in 1:nrow(AB_DIFF)){
AB_DIFF[i,2] = nrow(subdata[which(subdata$DIFF_Q == AB_DIFF[i,1]),])
}
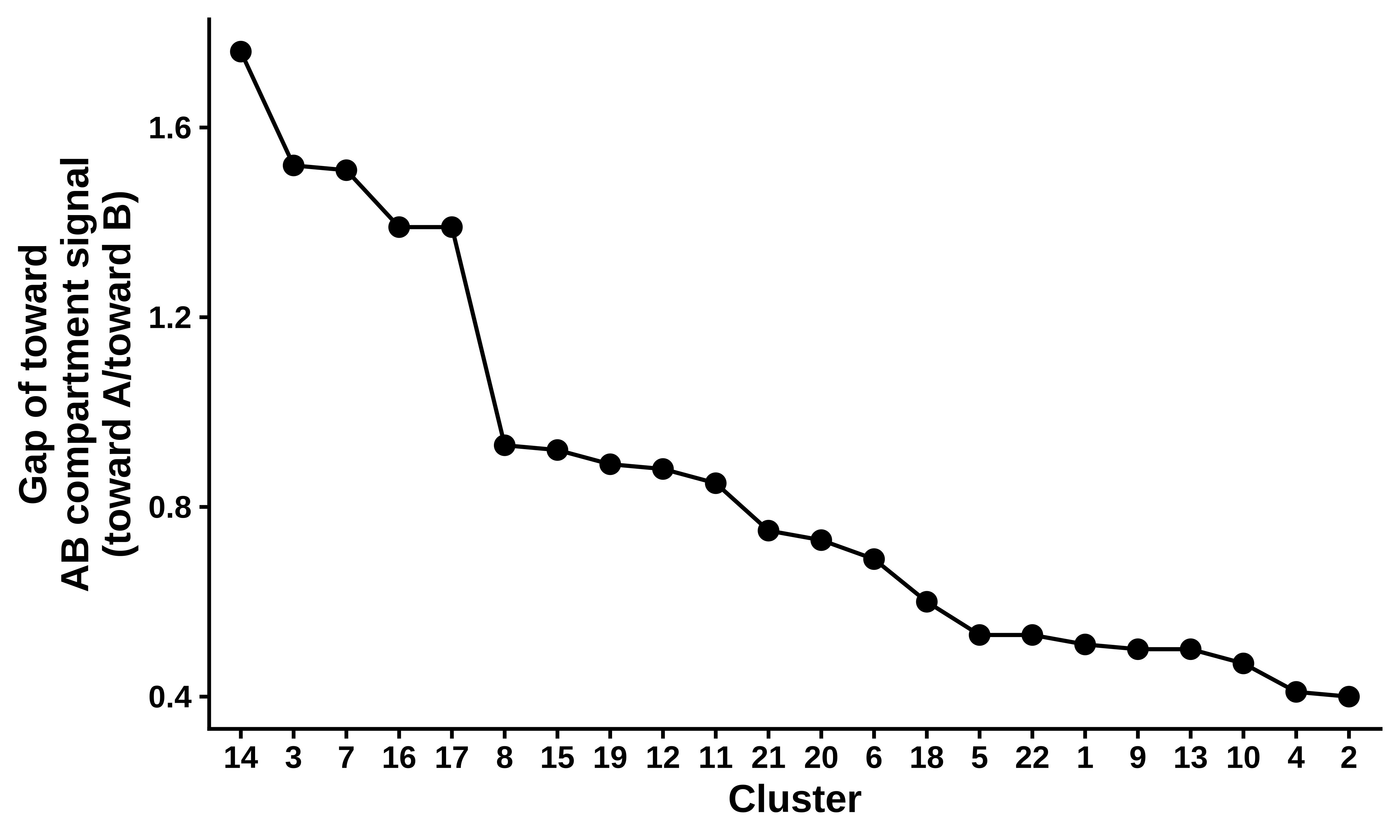
num_cluster_value = round(sum(AB_DIFF[7:10,2])/sum(AB_DIFF[1:5,2]),2)
DIFF_clusterDATA = as.data.frame(cbind(num_cluster,num_cluster_value))
for(s in 2:22){
num_cluster = s
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
AB_DIFF = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
AB_DIFF = as.data.frame(AB_DIFF)
colnames(AB_DIFF) = "AB_DIFF"
AB_DIFF$score = 0
for(i in 1:nrow(AB_DIFF)){
AB_DIFF[i,2] = nrow(subdata[which(subdata$DIFF_Q == AB_DIFF[i,1]),])
}
num_cluster_value = round(sum(AB_DIFF[7:10,2])/sum(AB_DIFF[1:5,2]),2)
DIFF_clusterDATA_frag = as.data.frame(cbind(num_cluster,num_cluster_value))
DIFF_clusterDATA = rbind(DIFF_clusterDATA,DIFF_clusterDATA_frag)
}
DIFF_clusterDATA = DIFF_clusterDATA[order(-DIFF_clusterDATA$num_cluster_value),]
DIFF_clustersorted = DIFF_clusterDATA[,1]
DIFF_clustersorted = as.data.frame(DIFF_clustersorted)
colnames(DIFF_clustersorted) = "cluster"
DIFF_clustersorted[,1] = as.factor(DIFF_clustersorted[,1])
DIFF_clusterDATA$sort = c(1:22)
DIFF_clusterDATA[,3] = as.factor(DIFF_clusterDATA[,3])
DIFF_clusterDATA$label = "DIFF"
options(repr.plot.width = 5, repr.plot.height = 3, repr.plot.res = 1000, repr.plot.pointsize = 40)
ggplot(DIFF_clusterDATA,aes(sort, y = num_cluster_value, group = "DIFF")) +
geom_line(color="black", size=0.5, alpha = 1) +
geom_point(color = "black", size = 2) +
theme_classic(base_size = 10) +
scale_x_discrete(labels = DIFF_clusterDATA[,1])+
ggtitle(NULL)+
xlab("Cluster") +
ylab("Gap of toward\nAB compartment signal\n(toward A/toward B)") +
theme(plot.title = element_text(face = "bold", hjust = 0.5, size = 25, color = "black"),
axis.text.x = element_text(face = "bold", size = 8),
axis.text.y = element_text(face = "bold", size = 8),
axis.title.x = element_text(face = "bold", size = 10, color = "black"),
axis.title.y = element_text(face = "bold", size = 10, color = "black"),
legend.position = "none",
legend.title = element_text(face = "bold", size = 10),
legend.text = element_text(face = "bold", size = 7))