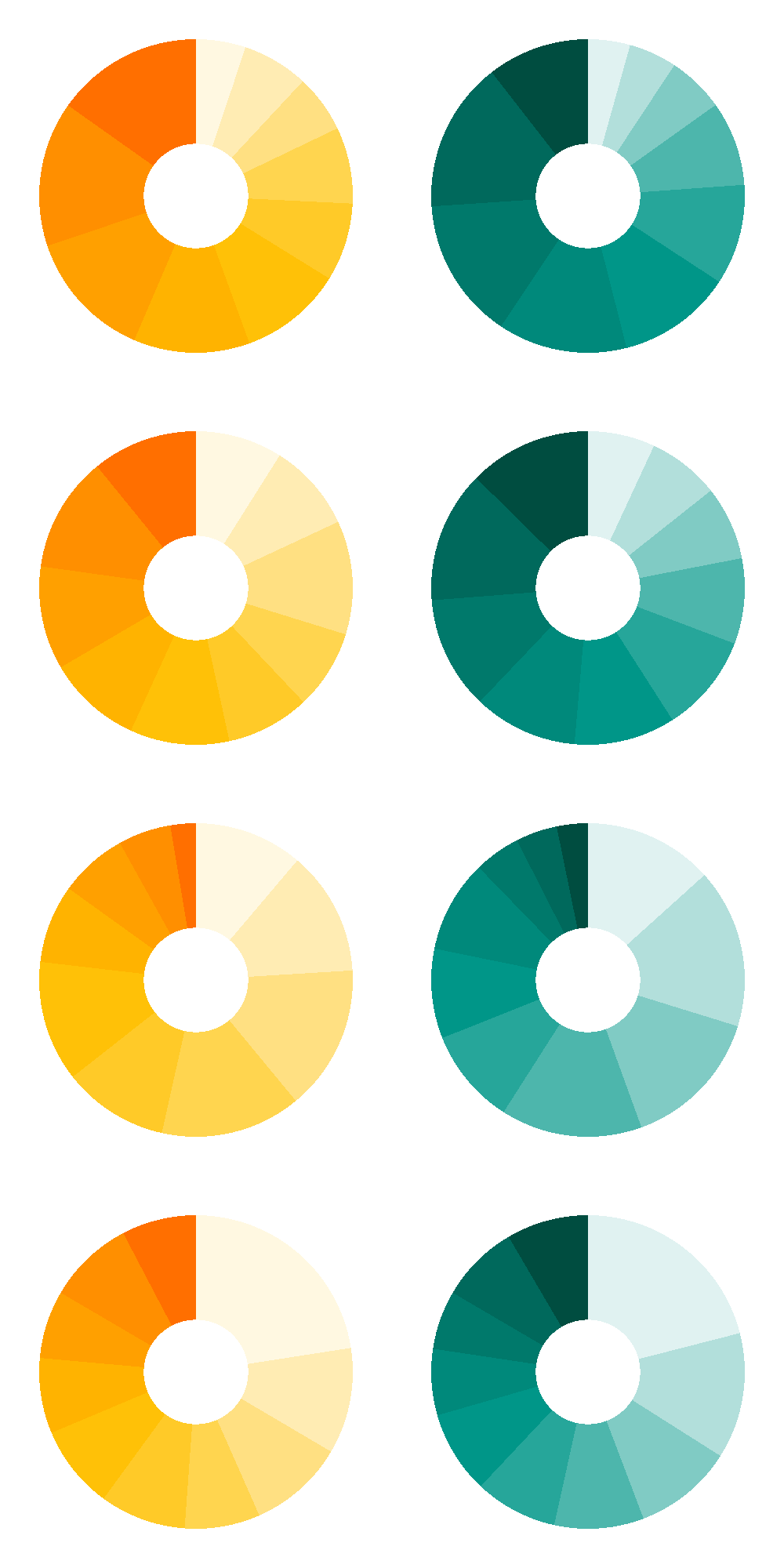5G
UMAP_DIFF_correlation_50_2_1_1 = as.data.frame(vroom("/Ets1/ProcessedData/UMAP_DIFF_correlation_50_2_1_1.txt", delim = "\t",show_col_types = FALSE))
UMAP_DIFF_correlation_50_2_1_1_fit = UMAP_DIFF_correlation_50_2_1_1[,1:2]
UMAP_DIFF_correlation_50_2_1_1_label = paste(UMAP_DIFF_correlation_50_2_1_1[,3],UMAP_DIFF_correlation_50_2_1_1[,4],UMAP_DIFF_correlation_50_2_1_1[,5])
UMAP_DIFF_correlation_50_2_1_1_label = as.data.frame(UMAP_DIFF_correlation_50_2_1_1_label)
colnames(UMAP_DIFF_correlation_50_2_1_1_label) = "label"
result <- kmeans(UMAP_DIFF_correlation_50_2_1_1_fit, centers = 22, iter.max = 10000, algorithm = "MacQueen")
UMAP_DIFF_correlation_50_2_1_1 = cbind(UMAP_DIFF_correlation_50_2_1_1,as.data.frame(result$cluster))
colnames(UMAP_DIFF_correlation_50_2_1_1)[ncol(UMAP_DIFF_correlation_50_2_1_1)] = "cluster"
cluster_Ets1_viewer = function(num_cluster){
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
Ets1_Q = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
Ets1_Q = as.data.frame(Ets1_Q)
colnames(Ets1_Q) = "Ets1_Q"
Ets1_Q$score = 0
for(i in 1:nrow(Ets1_Q)){
Ets1_Q[i,2] = nrow(subdata[which(subdata$Ets1_Q == Ets1_Q[i,1]),])
}
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
p300_Q = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
p300_Q = as.data.frame(p300_Q)
colnames(p300_Q) = "p300_Q"
p300_Q$score = 0
for(i in 1:nrow(p300_Q)){
p300_Q[i,2] = nrow(subdata[which(subdata$p300_Q == p300_Q[i,1]),])
}
ddata= cbind(Ets1_Q,p300_Q)
ddata$label = c("A","B","C","D","E","F","G","H","I","J")
options(repr.plot.width = 8, repr.plot.height = 8, repr.plot.res = 1000, repr.plot.pointsize = 10)
data <- data.frame(
category=ddata[,5],
count=ddata[,2])
data$fraction <- data$count / sum(data$count)
data$ymax <- cumsum(data$fraction)
data$ymin <- c(0, head(data$ymax, n=-1))
data$labelPosition <- (data$ymax + data$ymin) / 2
data$label <- paste0(data$category, "\n value: ", data$count)
nb.cols <- 19
mycolors <- colorRampPalette(brewer.pal(8, "Pastel2"))(nb.cols)
ggplot(data, aes(ymax=ymax, ymin=ymin, xmax=4, xmin=3, fill=category)) +
geom_rect() +
scale_color_manual(values=c("grey40"))+
scale_fill_manual(values=c("#FFF8E1","#FFECB3","#FFE082","#FFD54F","#FFCA28",
"#FFC107","#FFB300","#FFA000","#FF8F00","#FF6F00"))+
coord_polar(theta="y") +
coord_polar(theta="y") +
xlim(c(2.5, 4)) +
theme_void() +
scale_colour_hue(l = 20, c = 100)+
theme(legend.position = "none")
}
cluster_p300_viewer = function(num_cluster){
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
Ets1_Q = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
Ets1_Q = as.data.frame(Ets1_Q)
colnames(Ets1_Q) = "Ets1_Q"
Ets1_Q$score = 0
for(i in 1:nrow(Ets1_Q)){
Ets1_Q[i,2] = nrow(subdata[which(subdata$Ets1_Q == Ets1_Q[i,1]),])
}
subdata = as.data.frame(UMAP_DIFF_correlation_50_2_1_1[which(UMAP_DIFF_correlation_50_2_1_1$cluster == num_cluster),])
p300_Q = c("Q1","Q2","Q3","Q4","Q5","Q6","Q7","Q8","Q9","Q10")
p300_Q = as.data.frame(p300_Q)
colnames(p300_Q) = "p300_Q"
p300_Q$score = 0
for(i in 1:nrow(p300_Q)){
p300_Q[i,2] = nrow(subdata[which(subdata$p300_Q == p300_Q[i,1]),])
}
ddata= cbind(Ets1_Q,p300_Q)
ddata$label = c("A","B","C","D","E","F","G","H","I","J")
data <- data.frame(
category=ddata[,5],
count=ddata[,4])
data$fraction <- data$count / sum(data$count)
data$ymax <- cumsum(data$fraction)
data$ymin <- c(0, head(data$ymax, n=-1))
data$labelPosition <- (data$ymax + data$ymin) / 2
data$label <- paste0(data$category, "\n value: ", data$count)
nb.cols <- 19
mycolors <- colorRampPalette(brewer.pal(8, "Pastel2"))(nb.cols)
ggplot(data, aes(ymax=ymax, ymin=ymin, xmax=4, xmin=3, fill=category)) +
geom_rect() +
scale_color_manual(values=c("grey40"))+
scale_fill_manual(values=c("#E0F2F1","#B2DFDB","#80CBC4","#4DB6AC","#26A69A",
"#009688","#00897B","#00796B","#00695C","#004D40"))+
coord_polar(theta="y") +
coord_polar(theta="y") +
xlim(c(2.5, 4)) +
theme_void() +
scale_colour_hue(l = 20, c = 100)+
theme(legend.position = "none")
}
ppp=arrangeGrob(
cluster_Ets1_viewer(num_cluster=14),
cluster_p300_viewer(num_cluster=14),
cluster_Ets1_viewer(num_cluster=16),
cluster_p300_viewer(num_cluster=16),
cluster_Ets1_viewer(num_cluster=13),
cluster_p300_viewer(num_cluster=13),
cluster_Ets1_viewer(num_cluster=9),
cluster_p300_viewer(num_cluster=9),
ncol = 2,
nrow = 4)
options(repr.plot.width = 10, repr.plot.height = 20, repr.plot.res = 100, repr.plot.pointsize = 40)
grid.arrange(ppp)